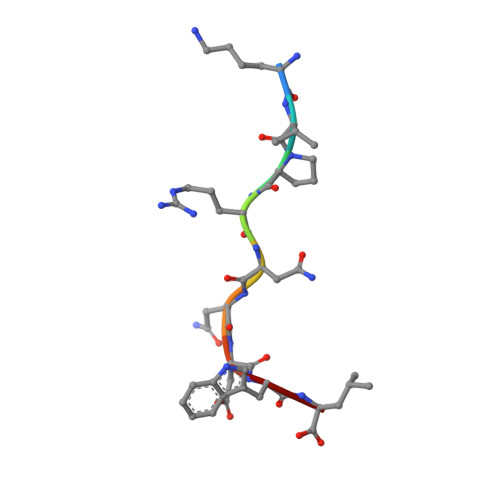
Design of agonistic altered peptides for the robust induction of CTL directed towards H-2Db in complex with the melanoma-associated epitope gp100.
van Stipdonk, M.J., Badia-Martinez, D., Sluijter, M., Offringa, R., van Hall, T., Achour, A.(2009) Cancer Res 69: 7784-7792
- PubMed: 19789338
- DOI: https://doi.org/10.1158/0008-5472.CAN-09-1724
- Primary Citation of Related Structures:
3CC5, 3CCH, 3CH1 - PubMed Abstract:
Immunogenicity of tumor-associated antigens (TAA) is often weak because many TAA are autoantigens for which the T-cell repertoire is sculpted by tolerance mechanisms. Substitutions at main anchor positions to increase the complementarity between the peptide and the MHC class I (MHC-I) binding cleft constitute a common procedure to improve binding capacity and immunogenicity of TAA. However, such alterations are tailored for each MHC-I allele and may recruit different CTL specificities through conformational changes in the targeted peptides. Comparative analysis of substituted melanoma-differentiation antigen gp100 in complex with H-2D(b) revealed that combined introduction of glycine and proline residues at the nonanchor positions 2 and 3, respectively, resulted in an agonistic altered peptide with dramatically enhanced binding affinity, stability, and immunogenicity of this TAA. Peptide vaccination using the p2Gp3P-altered peptide version of gp100 induced high frequencies of melanoma-specific CTL in the endogenous CD8+ repertoire. Crystal structure analysis of MHC/peptide complexes revealed that the conformation of the modified p2Gp3P-peptide was similar to the wild-type peptide, and indicated that this mimotope was stabilized through interactions between peptide residue p3P and the tyrosine residue Y159 that is conserved among most known MHC-I molecules throughout mammalian species. Our results may provide an alternative approach to enhance MHC stabilization capacity and immunogenicity of low-affinity peptides for induction of robust tumor-specific CTL.
- Departments of Immunohematology and Blood transfusion and Clinical Oncology, Leiden University Medical Center, Leiden, the Netherlands.
Organizational Affiliation: