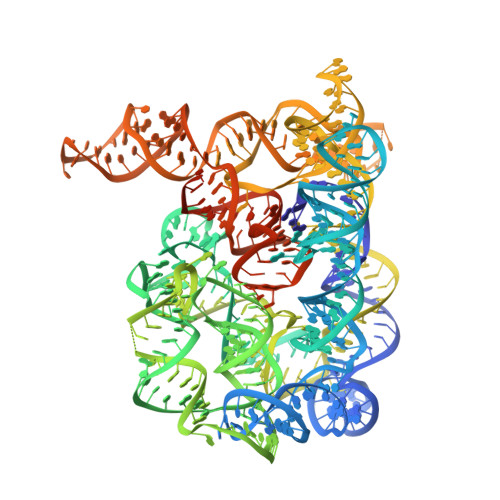
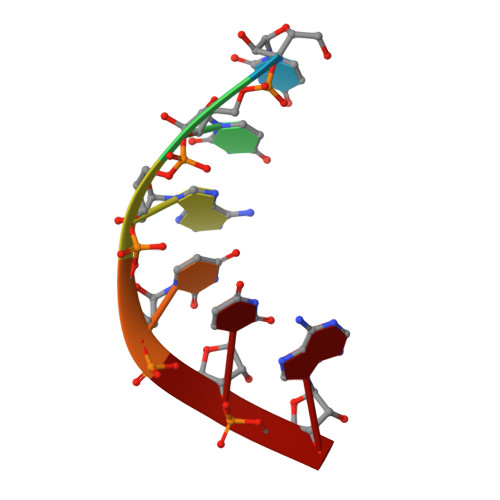
Structural basis for exon recognition by a group II intron.
Toor, N., Rajashankar, K., Keating, K.S., Pyle, A.M.(2008) Nat Struct Mol Biol 15: 1221-1222
- PubMed: 18953333
- DOI: https://doi.org/10.1038/nsmb.1509
- Primary Citation of Related Structures:
3EOG, 3EOH - PubMed Abstract:
Free group II introns are infectious retroelements that can bind and insert themselves into RNA and DNA molecules via reverse splicing. Here we report the 3.4-A crystal structure of a complex between an oligonucleotide target substrate and a group IIC intron, as well as the refined free intron structure. The structure of the complex reveals the conformation of motifs involved in exon recognition by group II introns.
- Department of Molecular Biophysics and Biochemistry, Yale University, 266 Whitney Avenue, New Haven, Connecticut 06520, USA.
Organizational Affiliation: