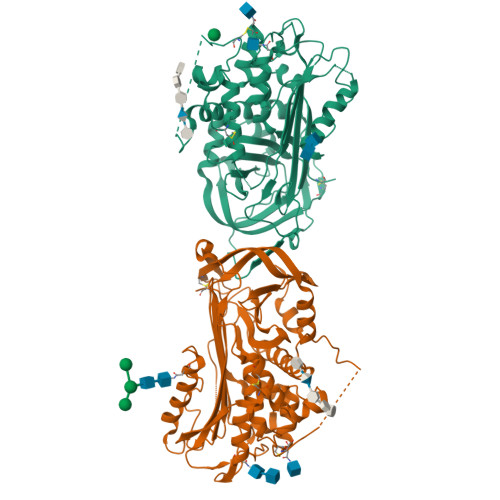
The critical role of hinge-region expulsion in the induced-fit heparin binding mechanism of antithrombin.
Langdown, J., Belzar, K.J., Savory, W.J., Baglin, T.P., Huntington, J.A.(2009) J Mol Biol 386: 1278-1289
- PubMed: 19452598
- DOI: https://doi.org/10.1016/j.jmb.2009.01.028
- Primary Citation of Related Structures:
3EVJ - PubMed Abstract:
Antithrombin (AT) is the most important inhibitor of coagulation proteases. Its activity is stimulated by glycosaminoglycans, such as heparin, through allosteric and template mechanisms. AT utilises an induced-fit mechanism to bind with high affinity to a pentasaccharide sequence found in about one-third of heparin chains. The conformational changes behind this mechanism have been characterised by several crystal structures of AT in the absence and in the presence of pentasaccharide. Pentasaccharide binding ultimately results in a conformational change that improves affinity by about 1000-fold. Crystal structures show several differences, including the expulsion of the hinge region of the reactive centre loop from beta-sheet A, which is known to be critical for the allosteric activation of AT. Here, we present data that reveal an energetically distinct intermediate on the path to full activation where the majority of conformational changes have already occurred. A crystal structure of this intermediate shows that the hinge region is in a native-like state in spite of having the pentasaccharide bound in the normal fashion. We engineered a disulfide bond to lock the hinge in its native position to determine the energetic contributions of the initial and final conformational events. Approximately 60% of the free-energy contribution of conformational change is provided by the final step of hinge-region expulsion and subsequent closure of the main beta-sheet A. A new analysis of the individual structural changes provides a plausible mechanism for propagation of conformational change from the heparin binding site to the remote hinge region in beta-sheet A.
Organizational Affiliation:
Department of Haematology, Cambridge Institute for Medical Research, University of Cambridge, Wellcome Trust/MRC Building, Hills Road, Cambridge CB2 0XY, UK.