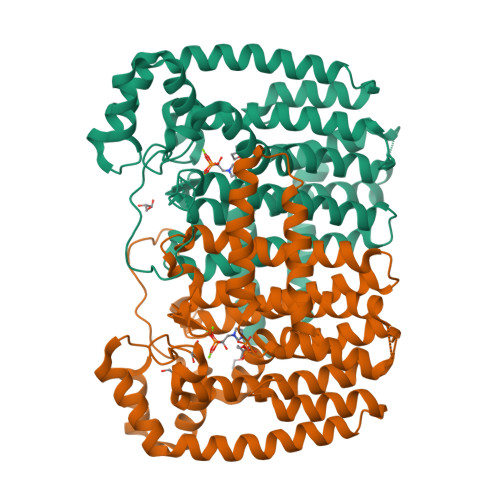
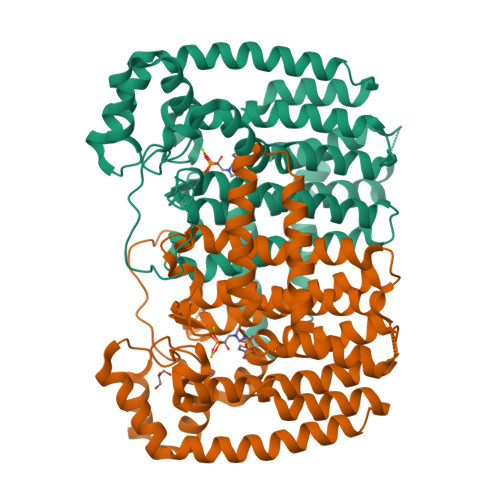
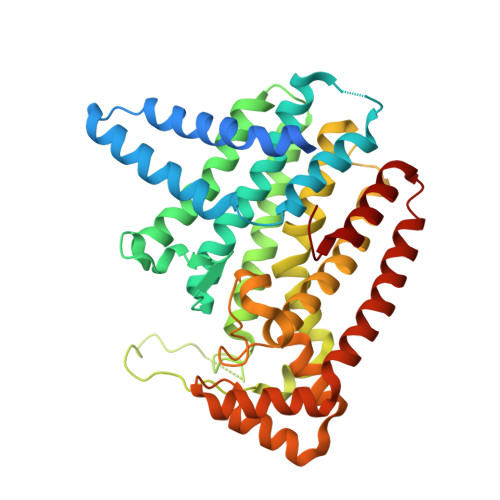
Crystal Structure of Plasmodium vivax geranylgeranylpyrophosphate synthase PVX_092040 with zoledronate and IPP bound
Wernimont, A.K., Lew, J., Zhao, Y., Kozieradzki, I., Cossar, D., Schapira, M., Bochkarev, A., Arrowsmith, C.H., Bountra, C., Weigelt, J., Edwards, A.M., Hui, R., Artz, J.D.To be published.