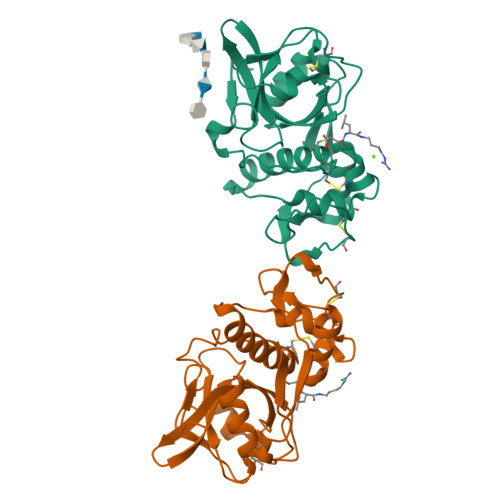
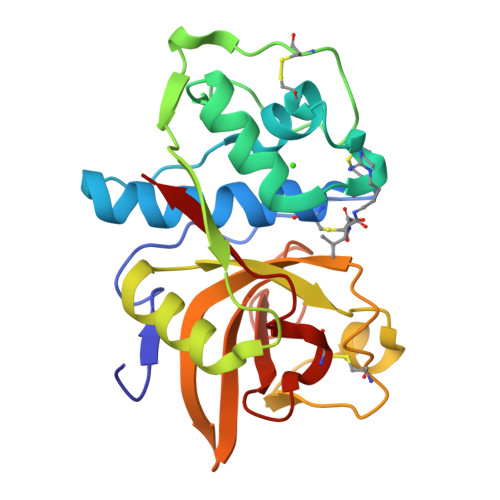
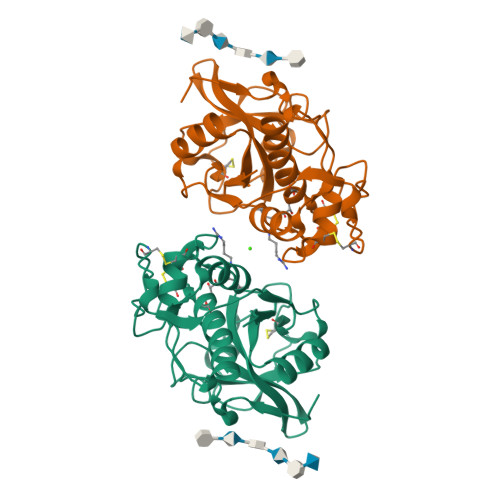
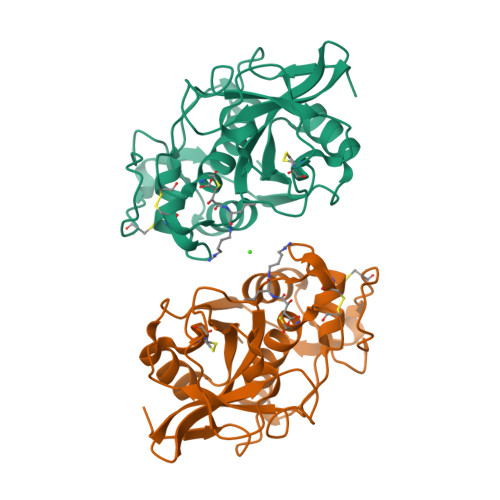
Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions.
Cherney, M.M., Lecaille, F., Kienitz, M., Nallaseth, F.S., Li, Z., James, M.N., Bromme, D.(2011) J Biol Chem 286: 8988-8998
- PubMed: 21193413
- DOI: https://doi.org/10.1074/jbc.M110.126706
- Primary Citation of Related Structures:
3H7D - PubMed Abstract:
In the presence of oligomeric chondroitin 4-sulfate (C4-S), cathepsin K (catK) forms a specific complex that was shown to be the source of the major collagenolytic activity in bone osteoclasts. C4-S forms multiple contacts with amino acid residues on the backside of the catK molecule that help to facilitate complex formation. As cathepsin L does not exhibit a significant collagenase activity in the presence or in the absence of C4-S, we substituted the C4-S interacting residues in catK with those of cathepsin L. Variants revealed altered collagenolytic activities with the largest inhibitory effect shown by the hexavariant M5. None of the variants showed a reduction in their gelatinolytic and peptidolytic activities when compared with wild-type catK, indicating no structural alteration within their active sites. However, the crystal structure of the M5 variant in the presence of oligomeric C4-S revealed a different binding of chondroitin 4-sulfate. C4-S is not continuously ordered as it is in the wild-type catK·C4-S complex. The orientation and the direction of the hexasaccharide on the catK surface have changed, so that the hexasaccharide is positioned between two symmetry-related molecules. Only one M5 variant molecule of the dimer that is present in the asymmetric unit interacts with C4-S. These substitutions have changed the mode of catK binding to C4-S and, as a result, have likely affected the collagenolytic potential of the variant. The data presented here support our hypothesis that distinct catK/C4-S interactions are necessary for the collagenolytic activity of the enzyme.
Organizational Affiliation:
Group in Protein Structure and Function, Department of Biochemistry, School of Molecular and Systems Medicine, University of Alberta, Edmonton, Alberta T6G 2H7, Canada.