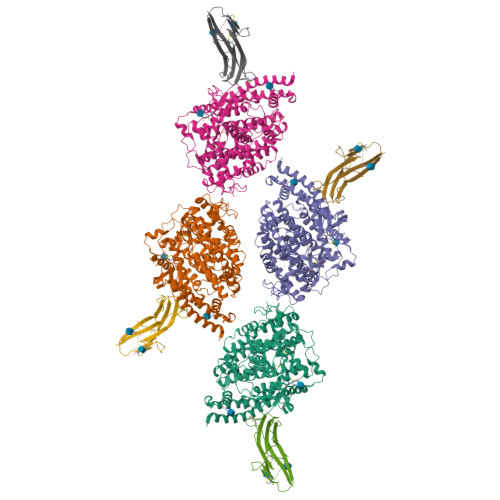
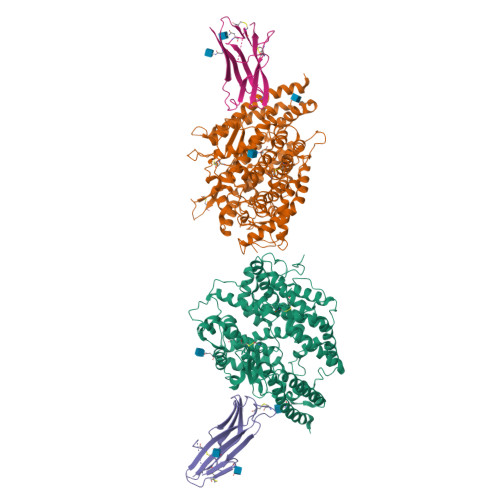
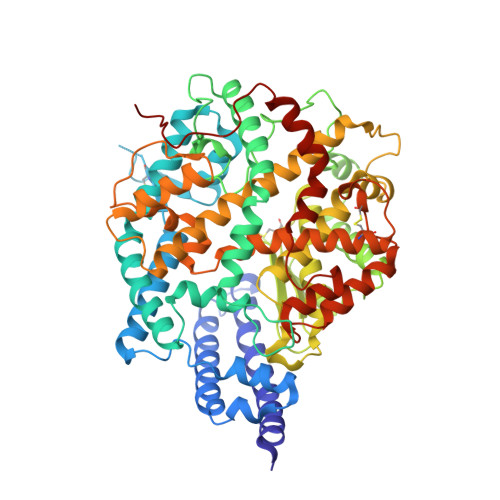
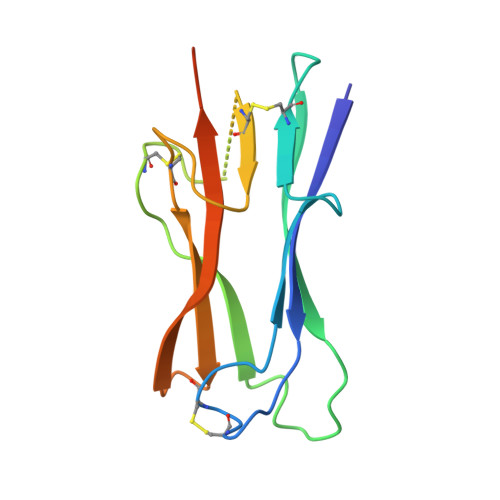
Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor.
Wu, K., Li, W., Peng, G., Li, F.(2009) Proc Natl Acad Sci U S A 106: 19970-19974
- PubMed: 19901337
- DOI: https://doi.org/10.1073/pnas.0908837106
- Primary Citation of Related Structures:
3KBH - PubMed Abstract:
NL63 coronavirus (NL63-CoV), a prevalent human respiratory virus, is the only group I coronavirus known to use angiotensin-converting enzyme 2 (ACE2) as its receptor. Incidentally, ACE2 is also used by group II SARS coronavirus (SARS-CoV). We investigated how different groups of coronaviruses recognize the same receptor, whereas homologous group I coronaviruses recognize different receptors. We determined the crystal structure of NL63-CoV spike protein receptor-binding domain (RBD) complexed with human ACE2. NL63-CoV RBD has a novel beta-sandwich core structure consisting of 2 layers of beta-sheets, presenting 3 discontinuous receptor-binding motifs (RBMs) to bind ACE2. NL63-CoV and SARS-CoV have no structural homology in RBD cores or RBMs; yet the 2 viruses recognize common ACE2 regions, largely because of a "virus-binding hotspot" on ACE2. Among group I coronaviruses, RBD cores are conserved but RBMs are variable, explaining how these viruses recognize different receptors. These results provide a structural basis for understanding viral evolution and virus-receptor interactions.
Organizational Affiliation:
Department of Pharmacology, University of Minnesota Medical School, Minneapolis, MN 55455, USA.