The IsdG-family of haem oxygenases degrades haem to a novel chromophore
Reniere, M.L., Ukpabi, G.N., Harry, S.R., Stec, D.F., Krull, R., Wright, D.W., Bachmann, B.O., Murphy, M.E.P., Skaar, E.P.(2010) Mol Microbiol 75: 1529-1538
- PubMed: 20180905
- DOI: https://doi.org/10.1111/j.1365-2958.2010.07076.x
- Primary Citation of Related Structures:
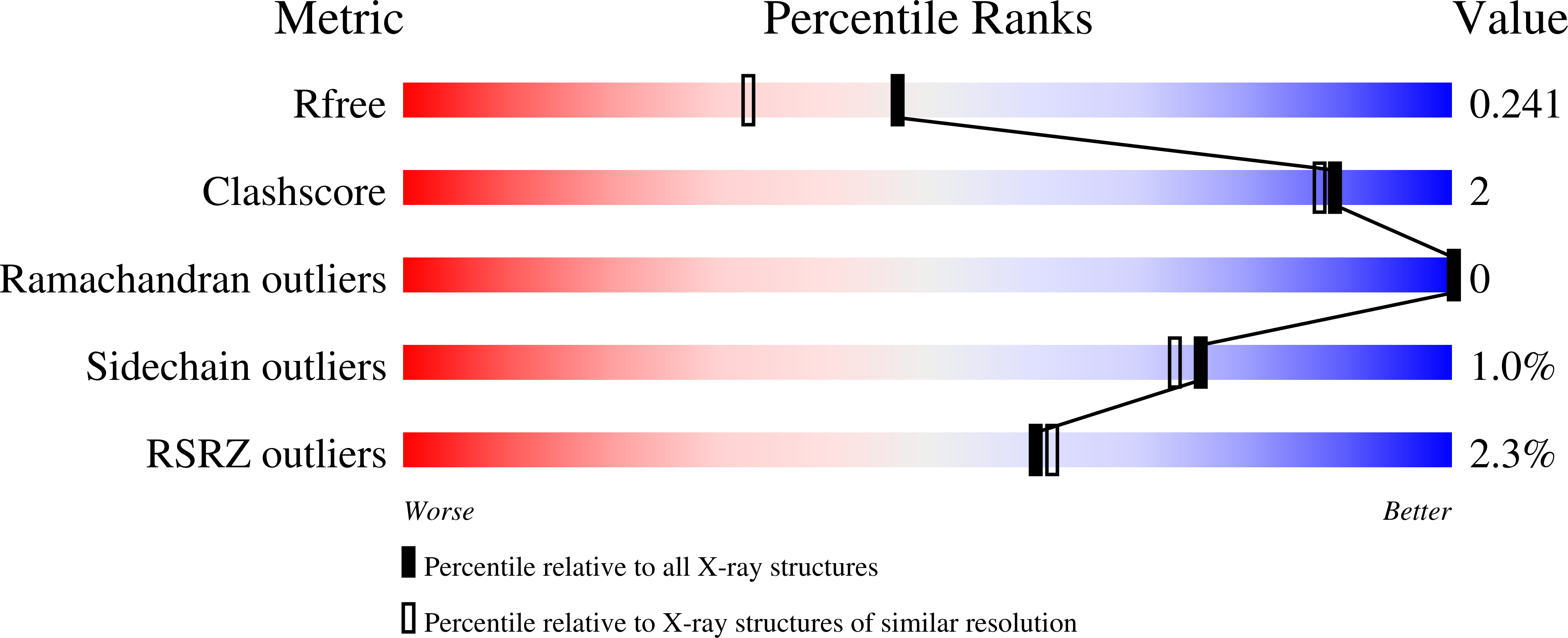
3LGM, 3LGN - PubMed Abstract:
Enzymatic haem catabolism by haem oxygenases is conserved from bacteria to humans and proceeds through a common mechanism leading to the formation of iron, carbon monoxide and biliverdin. The first members of a novel class of haem oxygenases were recently identified in Staphylococcus aureus (IsdG and IsdI) and were termed the IsdG-family of haem oxygenases. Enzymes of the IsdG-family form tertiary structures distinct from those of the canonical haem oxygenase family, suggesting that IsdG-family members degrade haem via a unique reaction mechanism. Herein we report that the IsdG-family of haem oxygenases degrade haem to the oxo-bilirubin chromophore staphylobilin. We also present the crystal structure of haem-bound IsdI in which haem ruffling and constrained binding of oxygen is consistent with cleavage of the porphyrin ring at the beta- or delta-meso carbons. Combined, these data establish that the IsdG-family of haem oxygenases degrades haem to a novel chromophore distinct from biliverdin.
Organizational Affiliation:
Department of Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, TN, USA.