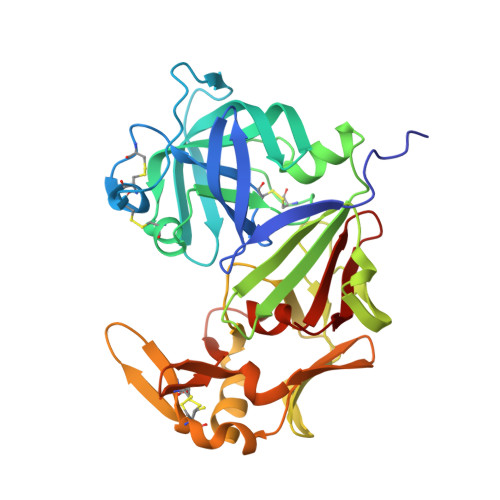
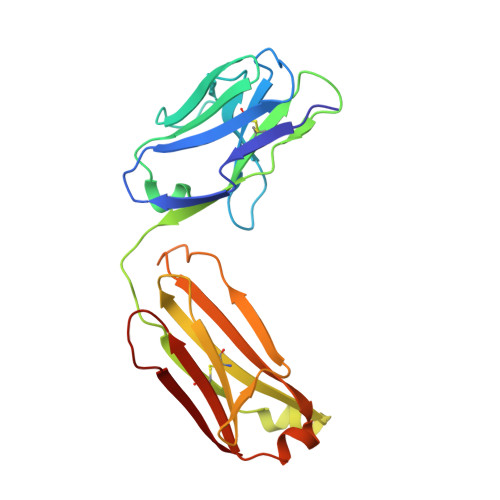
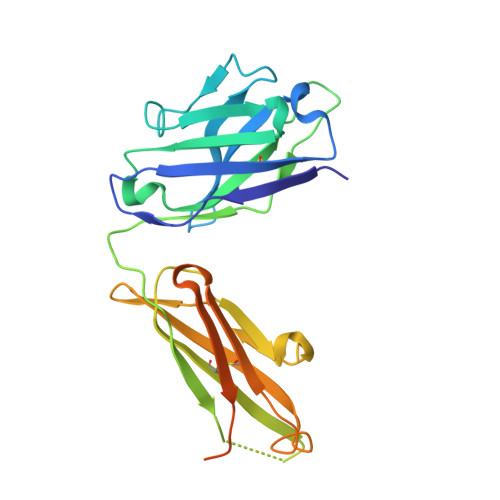
Mechanisms of allergen-antibody interaction of cockroach allergen Bla g 2 with monoclonal antibodies that inhibit IgE antibody binding.
Glesner, J., Wunschmann, S., Li, M., Gustchina, A., Wlodawer, A., Himly, M., Chapman, M.D., Pomes, A.(2011) PLoS One 6: e22223-e22223
- PubMed: 21789239
- DOI: https://doi.org/10.1371/journal.pone.0022223
- Primary Citation of Related Structures:
3LIZ - PubMed Abstract:
Cockroach allergy is strongly associated with asthma, and involves the production of IgE antibodies against inhaled allergens. Reports of conformational epitopes on inhaled allergens are limited. The conformational epitopes for two specific monoclonal antibodies (mAb) that interfere with IgE antibody binding were identified by X-ray crystallography on opposite sites of the quasi-symmetrical cockroach allergen Bla g 2. Mutational analysis of selected residues in both epitopes was performed based on the X-ray crystal structures of the allergen with mAb Fab/Fab' fragments, to investigate the structural basis of allergen-antibody interactions. The epitopes of Bla g 2 for the mAb 7C11 or 4C3 were mutated, and the mutants were analyzed by SDS-PAGE, circular dichroism, and/or mass spectrometry. Mutants were tested for mAb and IgE antibody binding by ELISA and fluorescent multiplex array. Single or multiple mutations of five residues from both epitopes resulted in almost complete loss of mAb binding, without affecting the overall folding of the allergen. Preventing glycosylation by mutation N268Q reduced IgE binding, indicating a role of carbohydrates in the interaction. Cation-π interactions, as well as electrostatic and hydrophobic interactions, were important for mAb and IgE antibody binding. Quantitative differences in the effects of mutations on IgE antibody binding were observed, suggesting heterogeneity in epitope recognition among cockroach allergic patients. Analysis by site-directed mutagenesis of epitopes identified by X-ray crystallography revealed an overlap between monoclonal and IgE antibody binding sites and provided insight into the B cell repertoire to Bla g 2 and the mechanisms of allergen-antibody recognition, including involvement of carbohydrates.
Organizational Affiliation:
INDOOR Biotechnologies, Inc, Charlottesville, Virginia, United States of America.