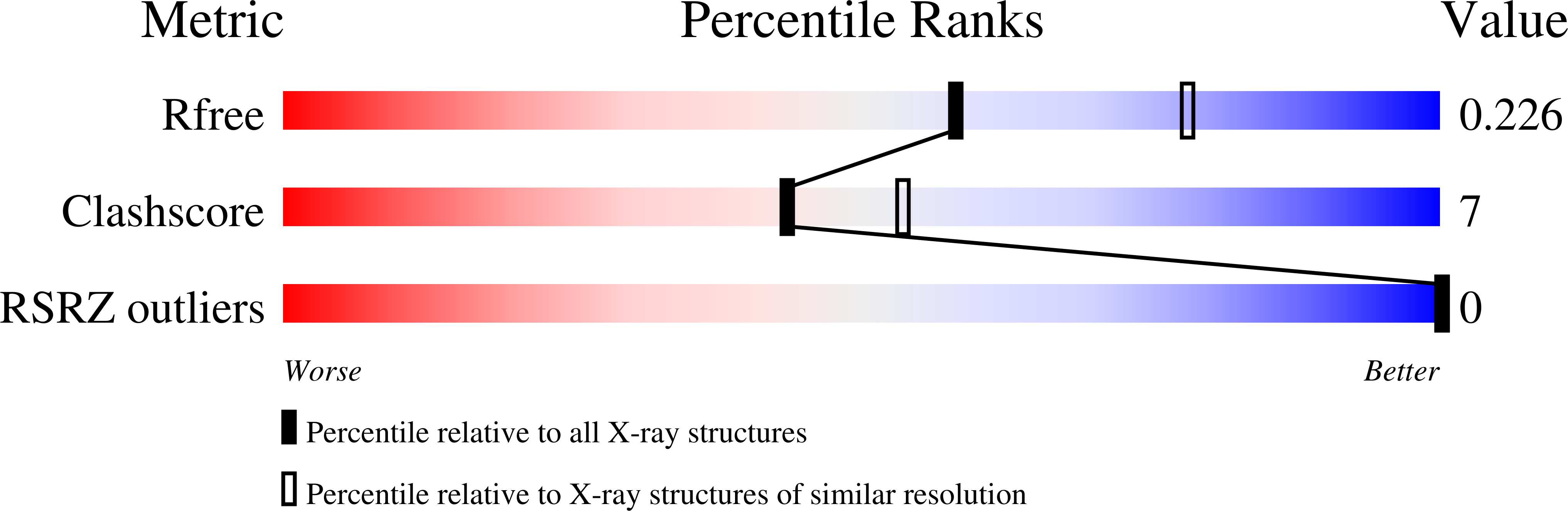X-Ray diffraction analyses of the natural isoquinoline alkaloids Berberine and Sanguinarine complexed with double helix DNA d(CGTACG)
Ferraroni, M., Bazzicalupi, C., Bilia, A.R., Gratteri, P.(2011) Chem Commun (Camb) 47: 4917-4919
- PubMed: 21431128
- DOI: https://doi.org/10.1039/c1cc10971e
- Primary Citation of Related Structures:
3NP6, 3NX5 - PubMed Abstract:
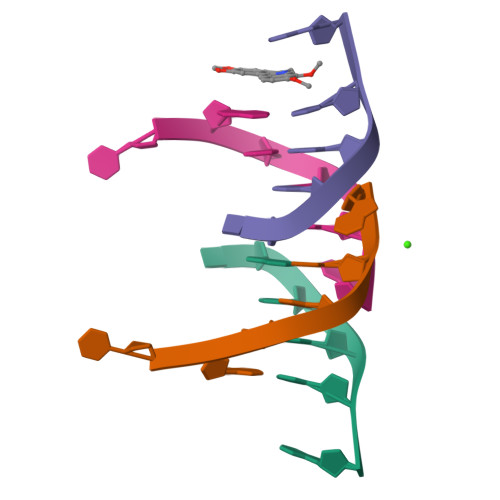
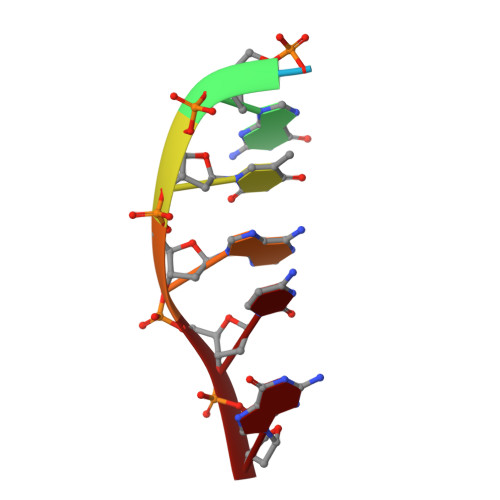
The first crystal structures of Berberine and Sanguinarine intercalated with a d(CGTACG)(2) DNA sequence were obtained by X-ray diffraction analysis at 2.3 Å resolution. Both drugs join the end of two "two-molecules" DNA units, stacked in a non-classic intercalation site formed by six bases. Sanguinarine interacts with d(CGTACG)(2) DNA in its iminium form.
Organizational Affiliation:
Department of Chemistry Ugo Schiff, University of Firenze, Via della Lastruccia 3-13, I-50019 Sesto Fiorentino, Firenze, Italy.