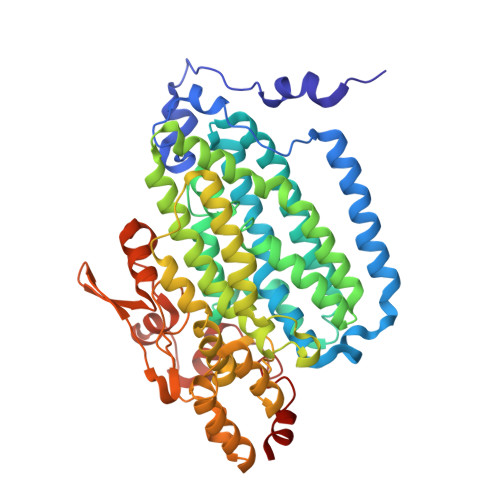
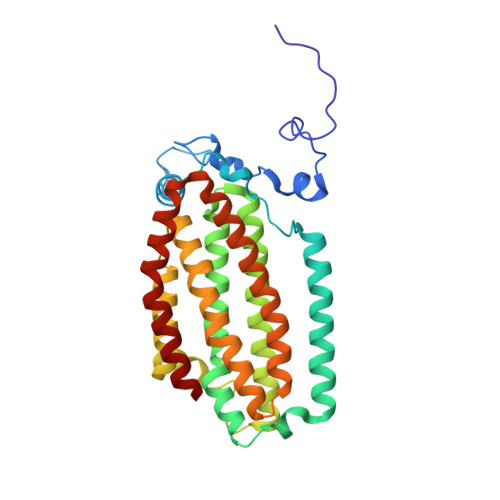
Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Bailey, L.J., Acheson, J.F., McCoy, J.G., Elsen, N.L., Phillips Jr., G.N., Fox, B.G.(2012) Biochemistry 51: 1101-1113
- PubMed: 22264099
- DOI: https://doi.org/10.1021/bi2018333
- Primary Citation of Related Structures:
3Q14, 3Q2A, 3Q3M, 3Q3N, 3Q3O, 3RI7, 3RMK - PubMed Abstract:
Crystal structures of toluene 4-monooxygenase hydroxylase in complex with reaction products and effector protein reveal active site interactions leading to regiospecificity. Complexes with phenolic products yield an asymmetric μ-phenoxo-bridged diiron center and a shift of diiron ligand E231 into a hydrogen bonding position with conserved T201. In contrast, complexes with inhibitors p-NH(2)-benzoate and p-Br-benzoate showed a μ-1,1 coordination of carboxylate oxygen between the iron atoms and only a partial shift in the position of E231. Among active site residues, F176 trapped the aromatic ring of products against a surface of the active site cavity formed by G103, E104 and A107, while F196 positioned the aromatic ring against this surface via a π-stacking interaction. The proximity of G103 and F176 to the para substituent of the substrate aromatic ring and the structure of G103L T4moHD suggest how changes in regiospecificity arise from mutations at G103. Although effector protein binding produced significant shifts in the positions of residues along the outer portion of the active site (T201, N202, and Q228) and in some iron ligands (E231 and E197), surprisingly minor shifts (<1 Å) were produced in F176, F196, and other interior residues of the active site. Likewise, products bound to the diiron center in either the presence or absence of effector protein did not significantly shift the position of the interior residues, suggesting that positioning of the cognate substrates will not be strongly influenced by effector protein binding. Thus, changes in product distributions in the absence of the effector protein are proposed to arise from differences in rates of chemical steps of the reaction relative to motion of substrates within the active site channel of the uncomplexed, less efficient enzyme, while structural changes in diiron ligand geometry associated with cycling between diferrous and diferric states are discussed for their potential contribution to product release.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison, Wisconsin 53706-1544, United States.