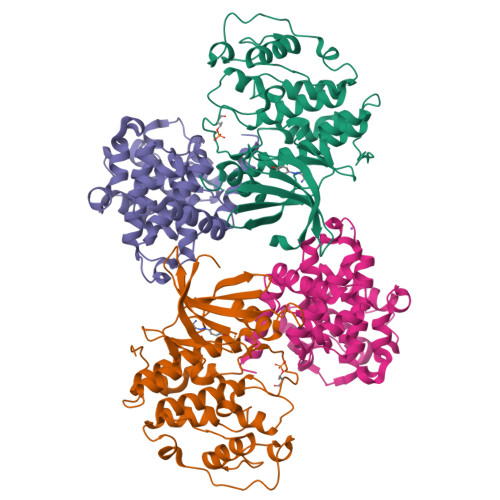
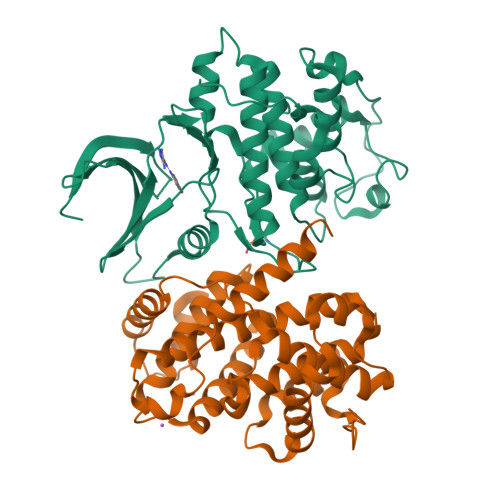
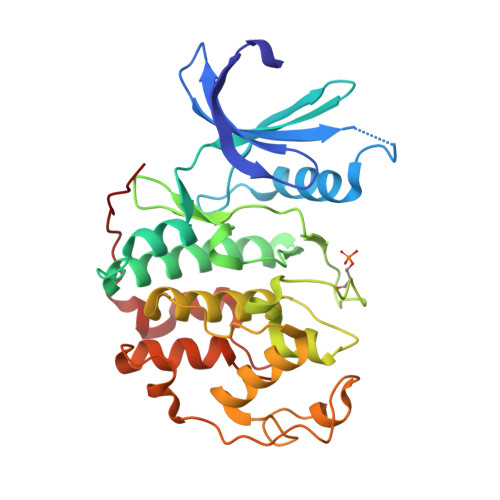
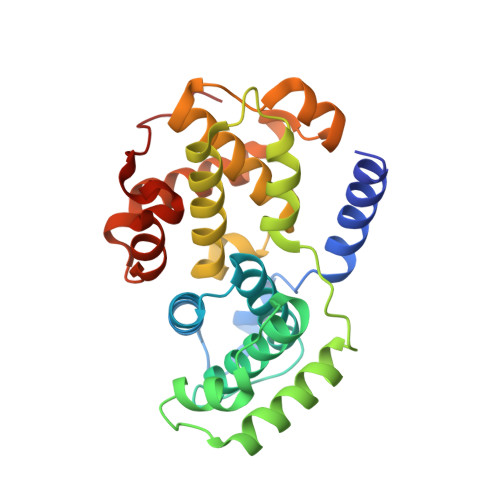
The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Baumli, S., Hole, A.J., Noble, M.E., Endicott, J.A.(2012) ACS Chem Biol 7: 811-816
- PubMed: 22292676
- DOI: https://doi.org/10.1021/cb2004516
- Primary Citation of Related Structures:
3TN8, 3TNH, 3TNI, 3TNW - PubMed Abstract:
CDK9 is the kinase of positive transcription elongation factor b and facilitates the transition of paused RNA polymerase II to processive transcription elongation. CDK9 is a validated target for the treatment of cancer, cardiac hypertrophy, and human immunodeficiency virus. Here we analyze different CDK9/cyclin T variants to identify a form of the complex amenable to use in inhibitor design. To demonstrate the utility of this system, we have determined the crystal structures of CDK9/cyclin T and CDK2/cyclin A bound to the CDK9-specific inhibitor CAN508. Comparison of the structures reveals CDK9-specific conformational changes and identifies a CDK9-specific hydrophobic pocket, adjacent to the αC-helix. By comparison with a previously published structure of CDK9/cyclin T/human immunodeficiency virus TAT we find that the CDK9 αC-helix has a degree of conformational variability that has the potential to be exploited for inhibitor design.
Organizational Affiliation:
Northern Institute for Cancer Research, Newcastle University, Framlington Place, Newcastle upon Tyne NE2 4HH, U.K. sonja.baumli@ncl.ac.uk