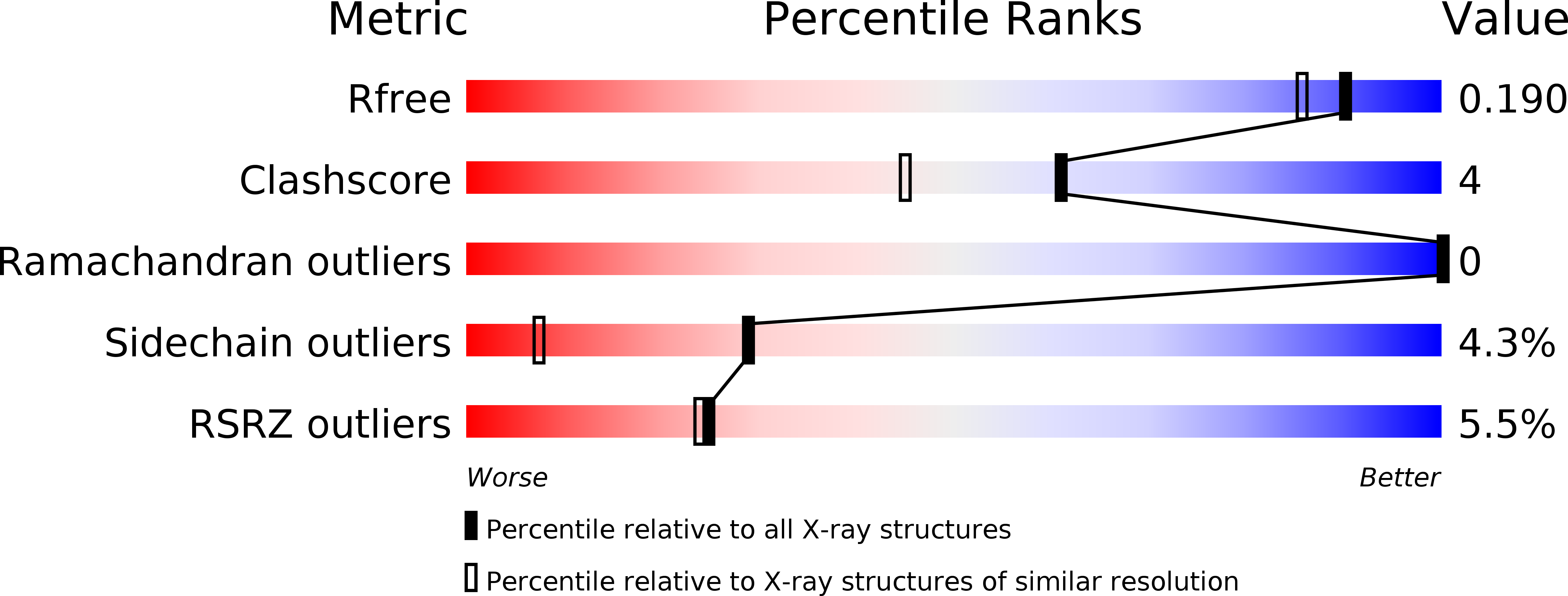
Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination.
Wang, Z., Michaud, G.A., Cheng, Z., Zhang, Y., Hinds, T.R., Fan, E., Cong, F., Xu, W.(2012) Genes Dev 26: 235-240
- PubMed: 22267412
- DOI: https://doi.org/10.1101/gad.182618.111
- Primary Citation of Related Structures:
3V3L - PubMed Abstract:
Protein poly(ADP-ribosyl)ation and ubiquitination are two key post-translational modifications regulating many biological processes. Through crystallographic and biochemical analysis, we show that the RNF146 WWE domain recognizes poly(ADP-ribose) (PAR) by interacting with iso-ADP-ribose (iso-ADPR), the smallest internal PAR structural unit containing the characteristic ribose-ribose glycosidic bond formed during poly(ADP-ribosyl)ation. The key iso-ADPR-binding residues we identified are highly conserved among WWE domains. Binding assays further demonstrate that PAR binding is a common function for the WWE domain family. Since many WWE domain-containing proteins are known E3 ubiquitin ligases, our results suggest that protein poly(ADP-ribosyl)ation may be a general mechanism to target proteins for ubiquitination.
Organizational Affiliation:
Department of Biological Structure, University of Washington, Seattle, Washington 98195, USA.