Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5,6]-fused bicyclic scaffolds
Okaniwa, M., Hirose, M., Imada, T., Ohashi, T., Hayashi, Y., Miyazaki, T., Arita, T., Yabuki, M., Kakoi, K., Kato, J., Takagi, T., Kawamoto, T., Yao, S., Sumita, A., Tsutsumi, S., Tottori, T., Oki, H., Sang, B.C., Yano, J., Aertgeerts, K., Yoshida, S., Ishikawa, T.(2012) J Med Chem 55: 3452-3478
- PubMed: 22376051
- DOI: https://doi.org/10.1021/jm300126x
- Primary Citation of Related Structures:
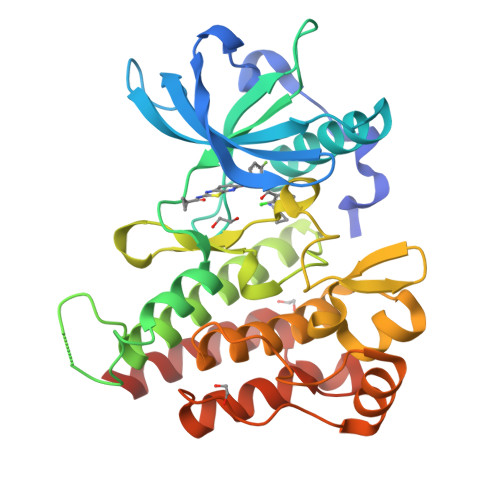
3VNT, 4DBN - PubMed Abstract:
To develop RAF/VEGFR2 inhibitors that bind to the inactive DFG-out conformation, we conducted structure-based drug design using the X-ray cocrystal structures of BRAF, starting from an imidazo[1,2-b]pyridazine derivative. We designed various [5,6]-fused bicyclic scaffolds (ring A, 1-6) possessing an anilide group that forms two hydrogen bond interactions with Cys532. Stabilizing the planarity of this anilide and the nitrogen atom on the six-membered ring of the scaffold was critical for enhancing BRAF inhibition. The selected [1,3]thiazolo[5,4-b]pyridine derivative 6d showed potent inhibitory activity in both BRAF and VEGFR2. Solid dispersion formulation of 6d (6d-SD) maximized its oral absorption in rats and showed significant suppression of ERK1/2 phosphorylation in an A375 melanoma xenograft model in rats by single administration. Tumor regression (T/C = -7.0%) in twice-daily repetitive studies at a dose of 50 mg/kg in rats confirmed that 6d is a promising RAF/VEGFR2 inhibitor showing potent anticancer activity.
Organizational Affiliation:
Pharmaceutical Research Division, Takeda Pharmaceutical Company Limited, 26-1, Muraoka-Higashi 2-Chome, Fujisawa, Kanagawa 251-8555, Japan. masanori.okaniwa@takeda.com