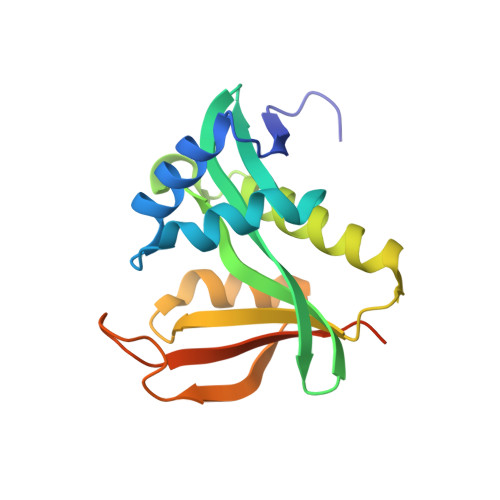
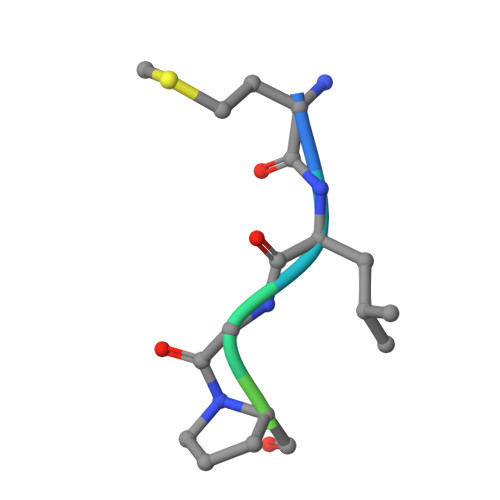
Structure of a Ternary Naa50p (NAT5/SAN) N-terminal Acetyltransferase Complex Reveals the Molecular Basis for Substrate-specific Acetylation.
Liszczak, G., Arnesen, T., Marmorstein, R.(2011) J Biol Chem 286: 37002-37010
- PubMed: 21900231
- DOI: https://doi.org/10.1074/jbc.M111.282863
- Primary Citation of Related Structures:
3TFY - PubMed Abstract:
The co-translational modification of N-terminal acetylation is ubiquitous among eukaryotes and has been reported to have a wide range of biological effects. The human N-terminal acetyltransferase (NAT) Naa50p (NAT5/SAN) acetylates the α-amino group of proteins containing an N-terminal methionine residue and is essential for proper sister chromatid cohesion and chromosome condensation. The elevated activity of NATs has also been correlated with cancer, making these enzymes attractive therapeutic targets. We report the x-ray crystal structure of Naa50p bound to a native substrate peptide fragment and CoA. We found that the peptide backbone of the substrate is anchored to the protein through a series of backbone hydrogen bonds with the first methionine residue specified through multiple van der Waals contacts, together creating an α-amino methionine-specific pocket. We also employed structure-based mutagenesis; the results support the importance of the α-amino methionine-specific pocket of Naa50p and are consistent with the proposal that conserved histidine and tyrosine residues play important catalytic roles. Superposition of the ternary Naa50p complex with the peptide-bound Gcn5 histone acetyltransferase revealed that the two enzymes share a Gcn5-related N-acetyltransferase fold but differ in their respective substrate-binding grooves such that Naa50p can accommodate only an α-amino substrate and not a side chain lysine substrate that is acetylated by lysine acetyltransferase enzymes such as Gcn5. The structure of the ternary Naa50p complex also provides the first molecular scaffold for the design of NAT-specific small molecule inhibitors with possible therapeutic applications.
Organizational Affiliation:
The Wistar Institute, Philadelphia, Pennsylvania 19104, USA.