Refined structure and solvent network of chicken gizzard G-actin DNase 1 complex at 1.8A resolution
Sasaki, K., Sakabe, K., Sakabe, N., Kondo, H., Shimomur, M.(1993) Acta Crystallogr A 49: C111-C112
Experimental Data Snapshot
(1993) Acta Crystallogr A 49: C111-C112
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin, gamma-enteric smooth muscle | 374 | Gallus gallus | Mutation(s): 0 EC: 3.6.4 | 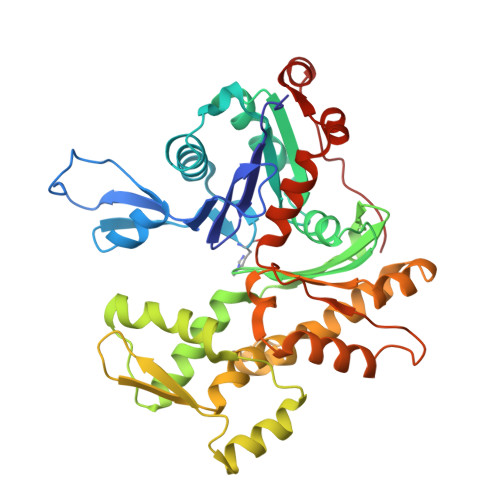 | |
UniProt | |||||
Find proteins for P63270 (Gallus gallus) Explore P63270 Go to UniProtKB: P63270 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63270 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Deoxyribonuclease-1 | 260 | Bos taurus | Mutation(s): 0 EC: 3.1.21.1 | 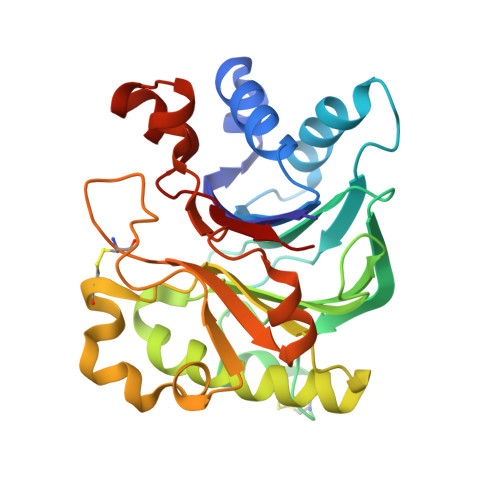 | |
UniProt | |||||
Find proteins for P00639 (Bos taurus) Explore P00639 Go to UniProtKB: P00639 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00639 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | C | 7 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G23450YU GlyCosmos: G23450YU GlyGen: G23450YU | |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | D [auth A] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| CA Query on CA | E [auth A], F [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| HIC Query on HIC | A | L-PEPTIDE LINKING | C7 H11 N3 O2 |  | HIS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42 | α = 90 |
| b = 225.3 | β = 90 |
| c = 77.4 | γ = 90 |
| Software Name | Purpose |
|---|---|
| WEIS | data scaling |
| BSS | data collection |
| X-PLOR | refinement |
| WEIS | data reduction |