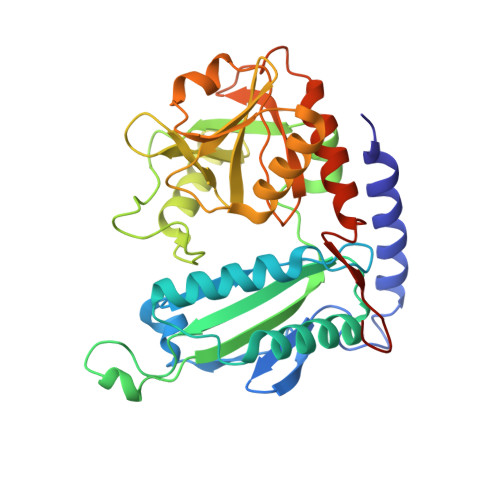
Crystal Structure of an (R)-Selective Omega-Transaminase from Aspergillus Terreus
Lyskowski, A., Gruber, C., Steinkellner, G., Schurmann, M., Schwab, H., Gruber, K., Steiner, K.(2014) PLoS One 9: 87350
- PubMed: 24498081
- DOI: https://doi.org/10.1371/journal.pone.0087350
- Primary Citation of Related Structures:
4CE5 - PubMed Abstract:
Chiral amines are important building blocks for the synthesis of pharmaceutical products, fine chemicals, and agrochemicals. ω-Transaminases are able to directly synthesize enantiopure chiral amines by catalysing the transfer of an amino group from a primary amino donor to a carbonyl acceptor with pyridoxal 5'-phosphate (PLP) as cofactor. In nature, (S)-selective amine transaminases are more abundant than the (R)-selective enzymes, and therefore more information concerning their structures is available. Here, we present the crystal structure of an (R)-ω-transaminase from Aspergillus terreus determined by X-ray crystallography at a resolution of 1.6 Å. The structure of the protein is a homodimer that displays the typical class IV fold of PLP-dependent aminotransferases. The PLP-cofactor observed in the structure is present in two states (i) covalently bound to the active site lysine (the internal aldimine form) and (ii) as substrate/product adduct (the external aldimine form) and free lysine. Docking studies revealed that (R)-transaminases follow a dual binding mode, in which the large binding pocket can harbour the bulky substituent of the amine or ketone substrate and the α-carboxylate of pyruvate or amino acids, and the small binding pocket accommodates the smaller substituent.
- ACIB GmbH, c/o TU Graz, Graz, Austria.
Organizational Affiliation: