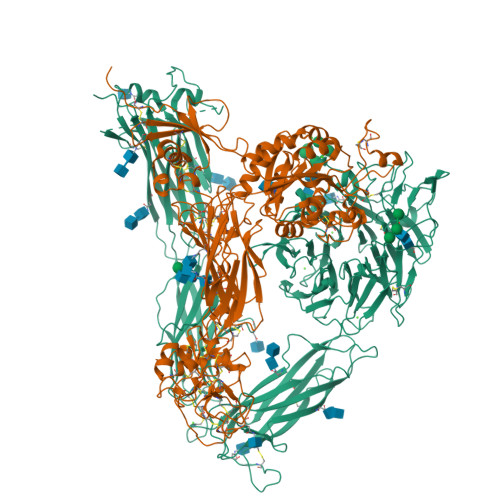
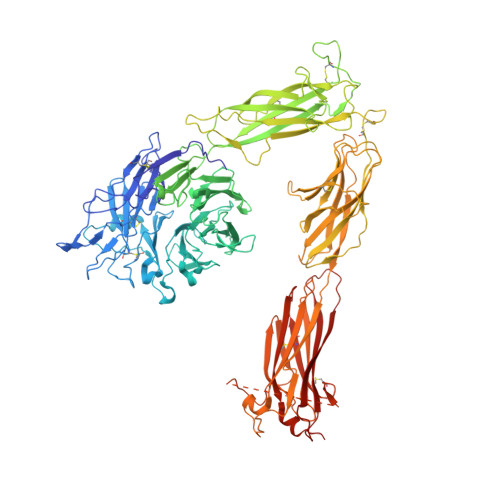
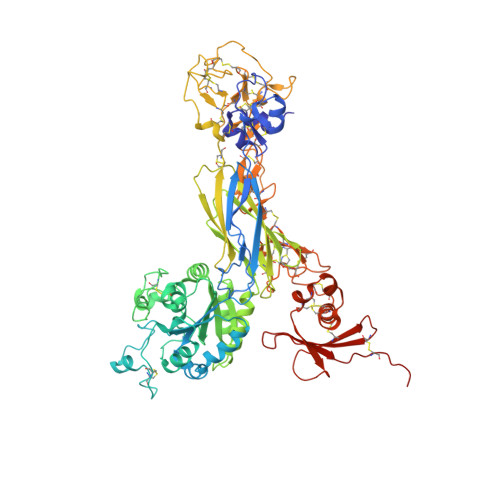
Alpha V Beta 3 Integrin Crystal Structures and their Functional Implications
Dong, X., Mi, L.Z., Zhu, J., Wang, W., Hu, P., Luo, B.H., Springer, T.A.(2012) Biochemistry 51: 8814-8828
- PubMed: 23106217
- DOI: https://doi.org/10.1021/bi300734n
- Primary Citation of Related Structures:
4G1E, 4G1M - PubMed Abstract:
Many questions about the significance of structural features of integrin α(V)β(3) with respect to its mechanism of activation remain. We have determined and re-refined crystal structures of the α(V)β(3) ectodomain linked to C-terminal coiled coils (α(V)β(3)-AB) and four transmembrane (TM) residues in each subunit (α(V)β(3)-1TM), respectively. The α(V) and β(3) subunits with four and eight extracellular domains, respectively, are bent at knees between the integrin headpiece and lower legs, and the headpiece has the closed, low-affinity conformation. The structures differ in the occupancy of three metal-binding sites in the βI domain. Occupancy appears to be related to the pH of crystallization, rather than to the physiologic regulation of ligand binding at the central, metal ion-dependent adhesion site. No electron density was observed for TM residues and much of the α(V) linker. α(V)β(3)-AB and α(V)β(3)-1TM demonstrate flexibility in the linker between their extracellular and TM domains, rather than the previously proposed rigid linkage. A previously postulated interface between the α(V) and β(3) subunits at their knees was also not supported, because it lacks high-quality density, required rebuilding in α(V)β(3)-1TM, and differed markedly between α(V)β(3)-1TM and α(V)β(3)-AB. Together with the variation in domain-domain orientation within their bent ectodomains between α(V)β(3)-AB and α(V)β(3)-1TM, these findings are compatible with the requirement for large structural changes, such as extension at the knees and headpiece opening, in conveying activation signals between the extracellular ligand-binding site and the cytoplasm.
Organizational Affiliation:
Immune Disease Institute, Children's Hospital Boston, and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 3 Blackfan Circle, Boston, Massachusetts 02115, United States.