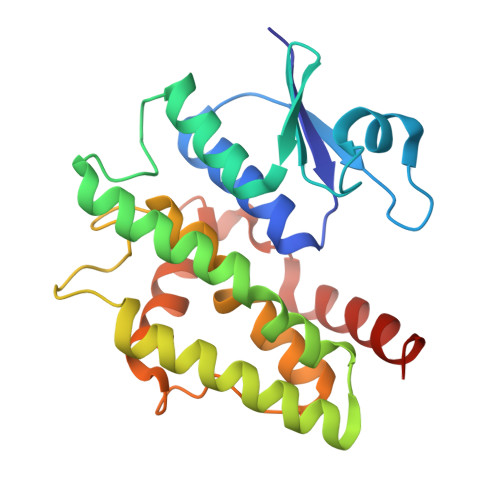
Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866
Kim, J., Toro, R., Bhosle, R., Al Obaidi, N.F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Washington, E., Scott Glenn, A., Chowdhury, S., Evans, B., Hammonds, J., Hillerich, B., Love, J., Seidel, R.D., Imker, H.J., Armstrong, R.N., Gerlt, J.A., Almo, S.C.To be published.