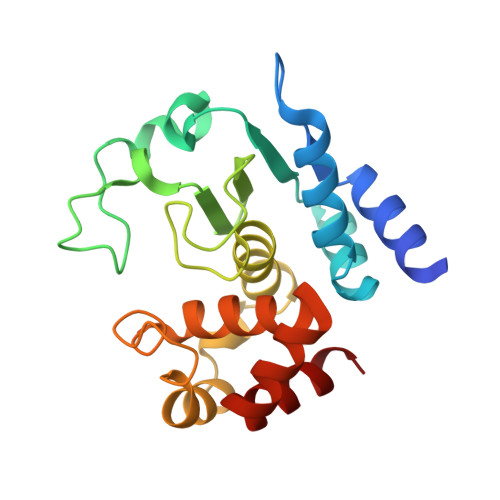
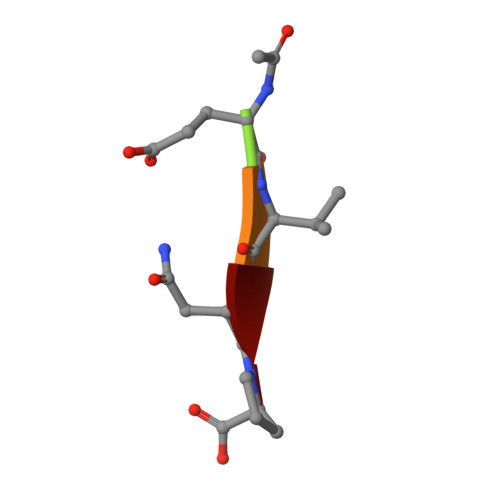
Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Schacherl, M., Pichlo, C., Neundorf, I., Baumann, U.(2015) Structure 23: 1632
- PubMed: 26211609
- DOI: https://doi.org/10.1016/j.str.2015.06.018
- Primary Citation of Related Structures:
5A0P, 5A0R, 5A0S, 5A0X - PubMed Abstract:
Clostridium difficile is a pathogenic bacterium causing gastrointestinal diseases from mild diarrhea to toxic megacolon. In common with other pathogenic bacteria, C. difficile secretes proteins involved in adhesion, colonization, and dissemination. The recently identified Zmp1 is an extracellular metalloprotease showing a unique specificity for Pro-Pro peptide bonds. The endogenous substrates of Zmp1 are two surface proteins implicated in adhesion of C. difficile to surface proteins of human cells. Thus, Zmp1 is believed to be involved in the regulation of the adhesion-motility balance of C. difficile. Here, we report crystal structures of Zmp1 from C. difficile in its unbound and peptide-bound forms. The structure analysis revealed a fold similar to Bacillus anthracis lethal factor. Crystal structures in the open and closed conformation of the S-loop shed light on the mode of binding of the substrate, and reveal important residues for substrate recognition and the strict specificity of Zmp1 for Pro-Pro peptide bonds.
Organizational Affiliation:
Institute of Biochemistry, University of Cologne, 50674 Cologne, Germany. Electronic address: magdalena.schacherl@uni-koeln.de.