2-Chloro-4-[[(1R,2R)-2-hydroxy-2-methyl-cyclopentyl]amino]-3-methyl-benzonitrile: A Transdermal Selective Androgen Receptor Modulator (SARM) for Muscle Atrophy.
Saeed, A., Vaught, G.M., Gavardinas, K., Matthews, D., Green, J.E., Losada, P.G., Bullock, H.A., Calvert, N.A., Patel, N.J., Sweetana, S.A., Krishnan, V., Henck, J.W., Luz, J.G., Wang, Y., Jadhav, P.(2016) J Med Chem 59: 750-755
- PubMed: 26683992
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01168
- Primary Citation of Related Structures:
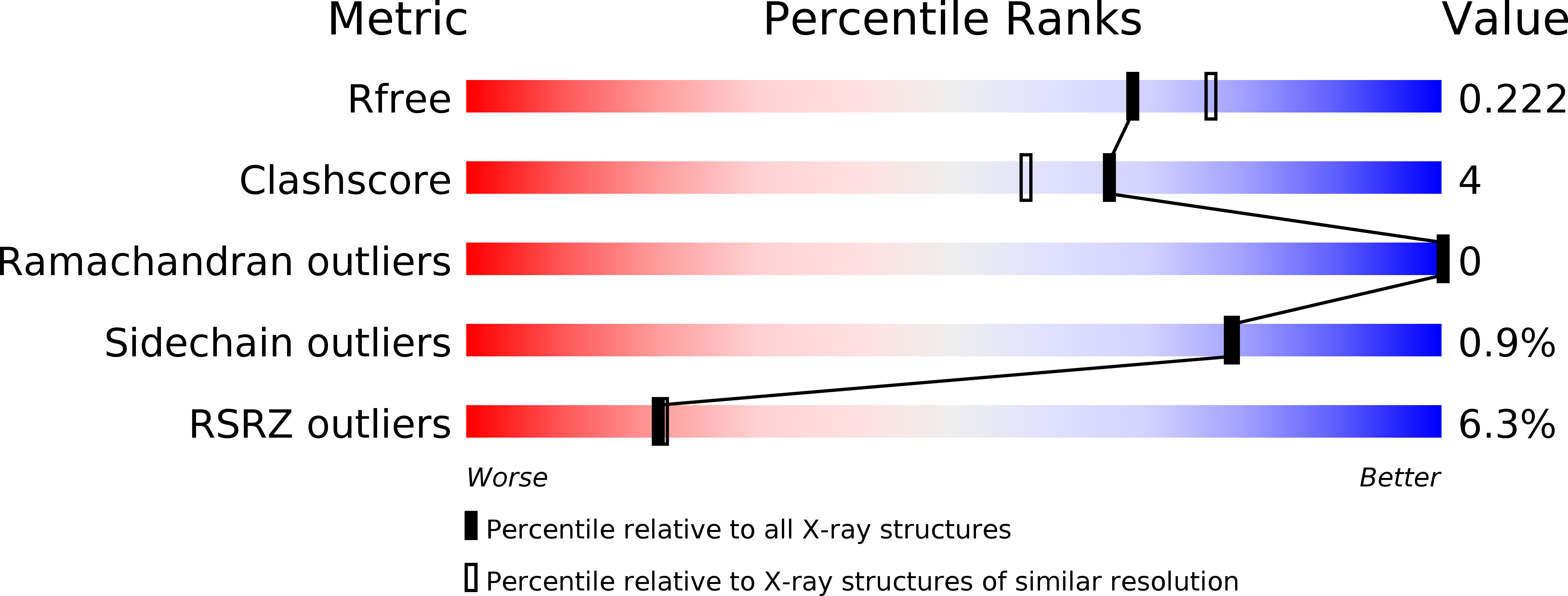
5CJ6 - PubMed Abstract:
A transdermal SARM has a potential to have therapeutic benefit through anabolic activity in muscle while sparing undesired effects of benign prostate hyperplasia (BPH) and liver-mediated decrease in HDL-C. 2-Chloro-4-[(2-hydroxy-2-methyl-cyclopentyl)amino]-3-methyl-benzonitrile 6 showed the desired muscle and prostate effects in a preclinical ORX rat model. Compound 6 had minimal effect on HDL-C levels in cynomolgus monkeys and showed human cadaver skin permeability, thus making it an effective tool for proof-of-concept studies in a clinical setting.
Organizational Affiliation:
Lilly Research Laboratories, Eli Lilly and Company , Lilly Corporate Center, Indianapolis, Indiana 46285, United States.