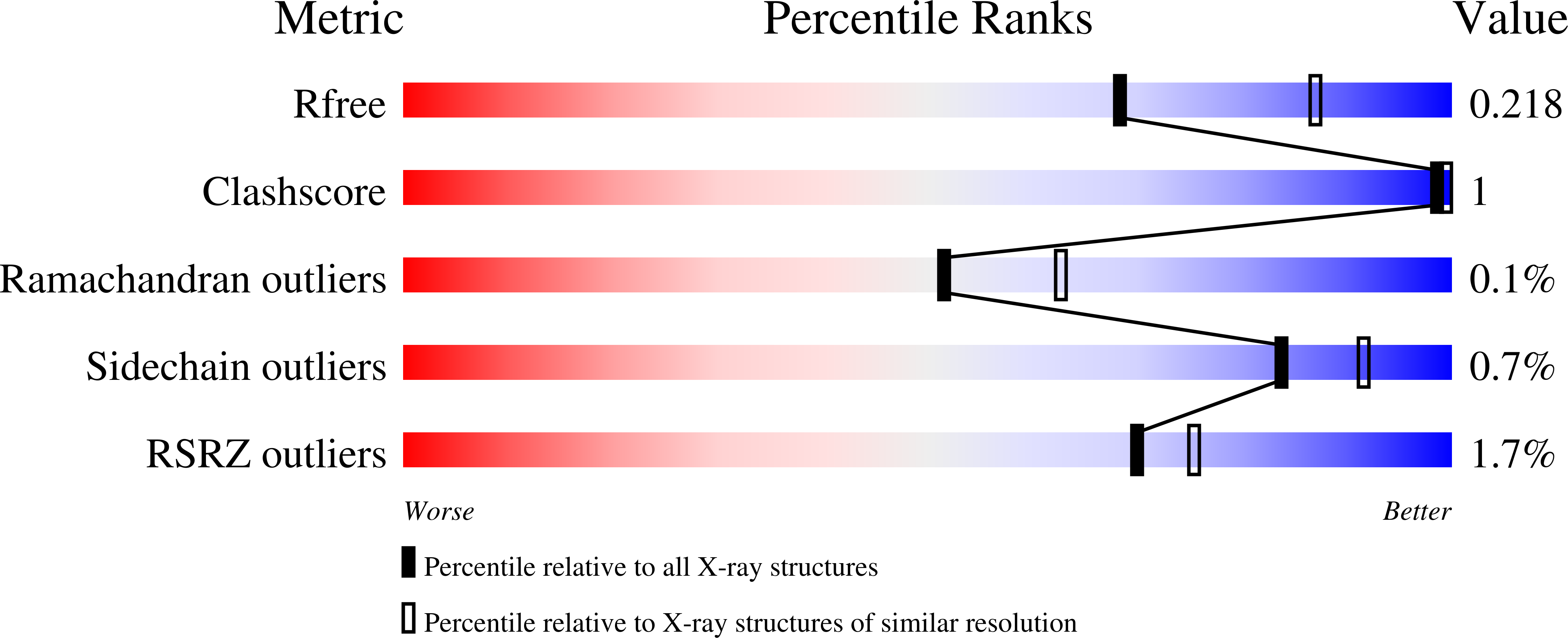
Structural determinants of nuclear export signal orientation in binding to exportin CRM1.
Fung, H.Y., Fu, S.C., Brautigam, C.A., Chook, Y.M.(2015) Elife 4
- PubMed: 26349033
- DOI: https://doi.org/10.7554/eLife.10034
- Primary Citation of Related Structures:
5DH9, 5DHA, 5DHF, 5DI9, 5DIF - PubMed Abstract:
The Chromosome Region of Maintenance 1 (CRM1) protein mediates nuclear export of hundreds of proteins through recognition of their nuclear export signals (NESs), which are highly variable in sequence and structure. The plasticity of the CRM1-NES interaction is not well understood, as there are many NES sequences that seem incompatible with structures of the NES-bound CRM1 groove. Crystal structures of CRM1 bound to two different NESs with unusual sequences showed the NES peptides binding the CRM1 groove in the opposite orientation (minus) to that of previously studied NESs (plus). Comparison of minus and plus NESs identified structural and sequence determinants for NES orientation. The binding of NESs to CRM1 in both orientations results in a large expansion in NES consensus patterns and therefore a corresponding expansion of potential NESs in the proteome.
Organizational Affiliation:
Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, United States.