Mutation of Tyr137 of the universal Escherichia coli fimbrial adhesin FimH relaxes the tyrosine gate prior to mannose binding.
Rabbani, S., Krammer, E.M., Roos, G., Zalewski, A., Preston, R., Eid, S., Zihlmann, P., Prevost, M., Lensink, M.F., Thompson, A., Ernst, B., Bouckaert, J.(2017) IUCrJ 4: 7-23
- PubMed: 28250938
- DOI: https://doi.org/10.1107/S2052252516016675
- Primary Citation of Related Structures:
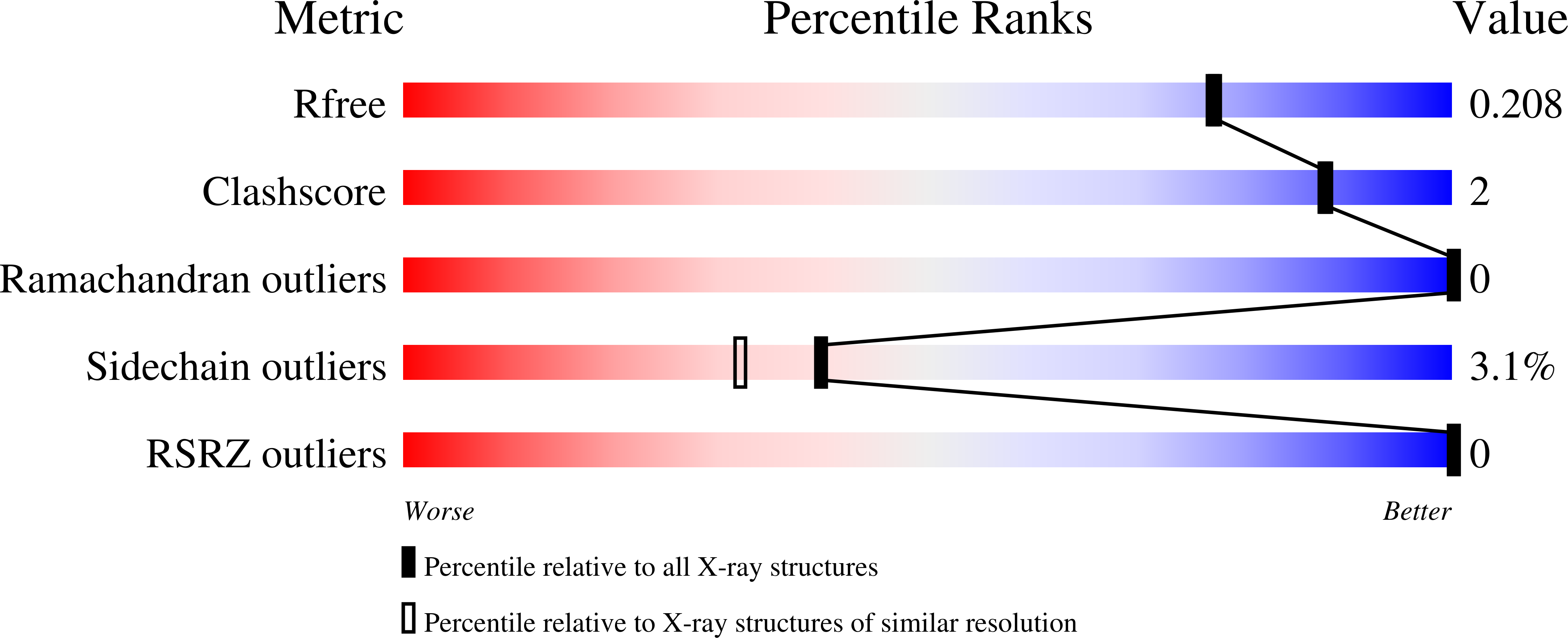
4CA4, 5FS5, 5FWR, 5FX3 - PubMed Abstract:
The most prevalent diseases manifested by Escherichia coli are acute and recurrent bladder infections and chronic inflammatory bowel diseases such as Crohn's disease. E. coli clinical isolates express the FimH adhesin, which consists of a mannose-specific lectin domain connected via a pilin domain to the tip of type 1 pili. Although the isolated FimH lectin domain has affinities in the nanomolar range for all high-mannosidic glycans, differentiation between these glycans is based on their capacity to form predominantly hydrophobic interactions within the tyrosine gate at the entrance to the binding pocket. In this study, novel crystal structures of tyrosine-gate mutants of FimH, ligand-free or in complex with heptyl α-d- O -mannopyranoside or 4-biphenyl α-d- O- mannopyranoside, are combined with quantum-mechanical calculations and molecular-dynamics simulations. In the Y48A FimH crystal structure, a large increase in the dynamics of the alkyl chain of heptyl α-d- O -mannopyranoside attempts to compensate for the absence of the aromatic ring; however, the highly energetic and stringent mannose-binding pocket of wild-type FimH is largely maintained. The Y137A mutation, on the other hand, is the most detrimental to FimH affinity and specificity: (i) in the absence of ligand the FimH C-terminal residue Thr158 intrudes into the mannose-binding pocket and (ii) ethylenediaminetetraacetic acid interacts strongly with Glu50, Thr53 and Asn136, in spite of multiple dialysis and purification steps. Upon mutation, pre-ligand-binding relaxation of the backbone dihedral angles at position 137 in the tyrosine gate and their coupling to Tyr48 via the interiorly located Ile52 form the basis of the loss of affinity of the FimH adhesin in the Y137A mutant.
Organizational Affiliation:
Institute of Molecular Pharmacy, Pharmacenter, University of Basel , Klingelbergstrasse 50-70, CH-4056 Basel, Switzerland.