Signaling States of a Short Blue-Light Photoreceptor Protein PpSB1-LOV Revealed from Crystal Structures and Solution NMR Spectroscopy.
Rollen, K., Granzin, J., Panwalkar, V., Arinkin, V., Rani, R., Hartmann, R., Krauss, U., Jaeger, K.E., Willbold, D., Batra-Safferling, R.(2016) J Mol Biol 428: 3721-3736
- PubMed: 27291287
- DOI: https://doi.org/10.1016/j.jmb.2016.05.027
- Primary Citation of Related Structures:
5J3W, 5J4E - PubMed Abstract:
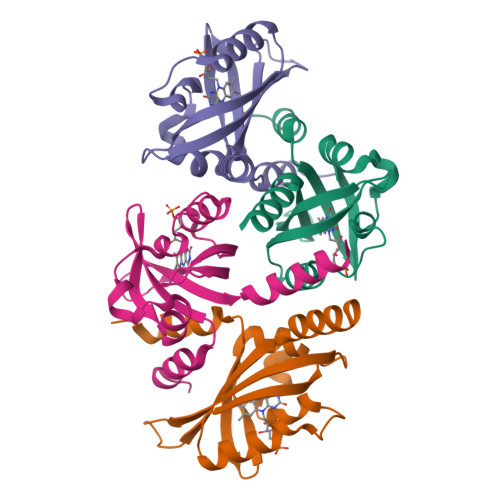
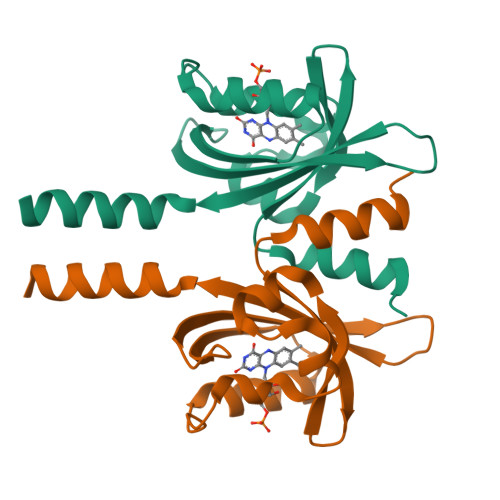
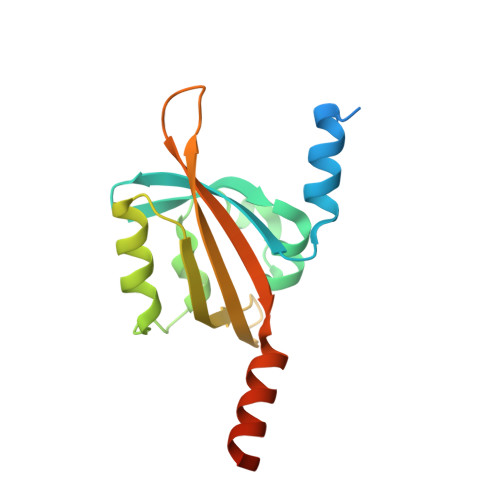
Light-Oxygen-Voltage (LOV) domains represent the photo-responsive domains of various blue-light photoreceptor proteins and are widely distributed in plants, algae, fungi, and bacteria. Here, we report the dark-state crystal structure of PpSB1-LOV, a slow-reverting short LOV protein from Pseudomonas putida that is remarkably different from our previously published "fully light-adapted" structure [1]. A direct comparison of the two structures provides insight into the light-activated signaling mechanism. Major structural differences involve a~11Å movement of the C terminus in helix Jα, ~4Å movement of Hβ-Iβ loop, disruption of hydrogen bonds in the dimer interface, and a~29° rotation of chain-B relative to chain-A as compared to the light-state dimer. Both crystal structures and solution NMR data are suggestive of the key roles of a conserved glutamine Q116 and the N-cap region consisting of A'α-Aβ loop and the A'α helix in controlling the light-activated conformational changes. The activation mechanism proposed here for the PpSB1-LOV supports a rotary switch mechanism and provides insights into the signal propagation mechanism in naturally existing and artificial LOV-based, two-component systems and regulators.
Organizational Affiliation:
Institute of Complex Systems, ICS-6: Structural Biochemistry, Forschungszentrum Jülich, 52425 Jülich, Germany.