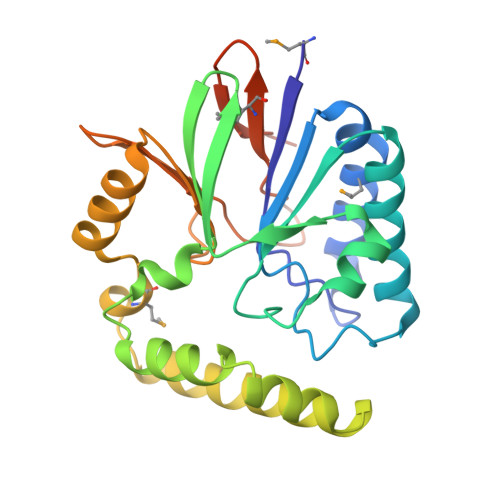
Structure of the essential Haemophilus influenzae UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis.
Cho, J., Lee, C.J., Zhao, J., Young, H.E., Zhou, P.(2016) Nat Microbiol 1: 16154-16154
- PubMed: 27780190
- DOI: https://doi.org/10.1038/nmicrobiol.2016.154
- Primary Citation of Related Structures:
5K8K - PubMed Abstract:
In most Gram-negative pathogens, the hydrolysis of UDP-2,3-diacylglucosamine to generate lipid X in lipid A biosynthesis is catalysed by the membrane-associated enzyme LpxH. We report the crystal structure of LpxH in complex with its product, lipid X, unveiling a unique insertion lid above the conserved architecture of calcineurin-like phosphoesterases. This structure reveals elaborate interactions surrounding lipid X and provides molecular insights into the substrate selectivity, catalysis and inhibition of LpxH.
- Department of Biochemistry, Duke University Medical Center, Research Drive, Durham, North Carolina 27710, USA.
Organizational Affiliation: