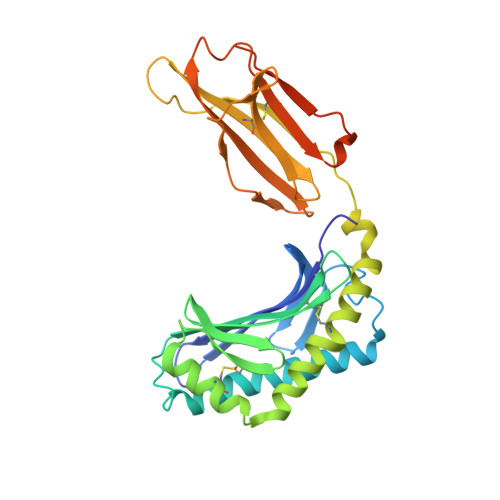
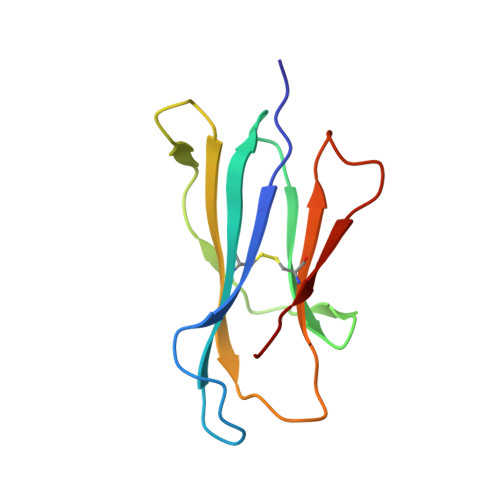
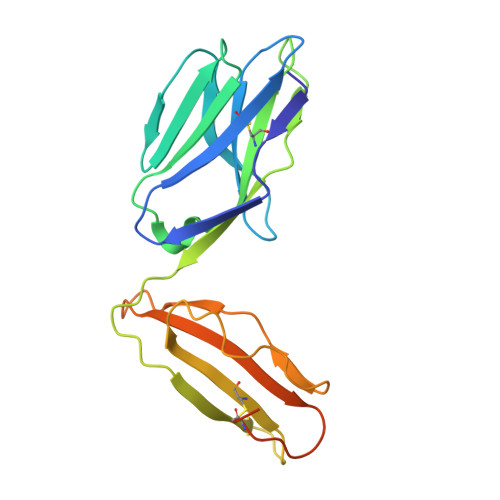
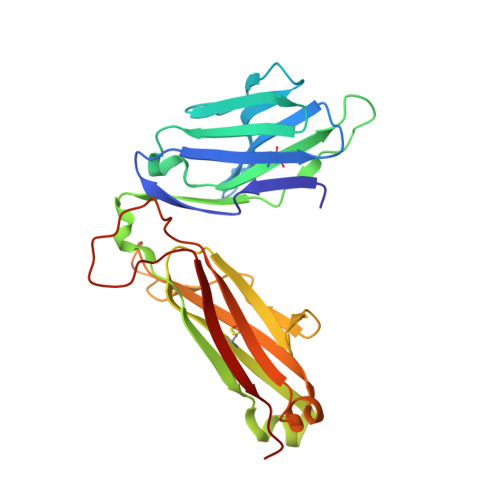
T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Gras, S., Van Rhijn, I., Shahine, A., Cheng, T.Y., Bhati, M., Tan, L.L., Halim, H., Tuttle, K.D., Gapin, L., Le Nours, J., Moody, D.B., Rossjohn, J.(2016) Nat Commun 7: 13257-13257
- PubMed: 27807341
- DOI: https://doi.org/10.1038/ncomms13257
- Primary Citation of Related Structures:
5L2J, 5L2K - PubMed Abstract:
CD1 proteins present microbial lipids to T cells. Germline-encoded mycolyl lipid-reactive (GEM) T cells with conserved αβ T cell receptors (TCRs) recognize CD1b presenting mycobacterial mycolates. As the molecular basis underpinning TCR recognition of CD1b remains unknown, here we determine the structure of a GEM TCR bound to CD1b presenting glucose-6-O-monomycolate (GMM). The GEM TCR docks centrally above CD1b, whereby the conserved TCR α-chain extensively contacts CD1b and GMM. Through mutagenesis and study of T cells from tuberculosis patients, we identify a consensus CD1b footprint of TCRs present among GEM T cells. Using both the TCR α- and β-chains as tweezers to surround and grip the glucose moiety of GMM, GEM TCRs create a highly specific mechanism for recognizing this mycobacterial glycolipid.
Organizational Affiliation:
Infection and Immunity Program, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.