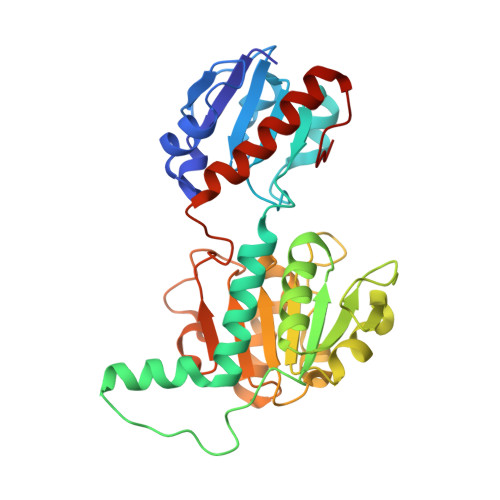
Potassium binding by carbonyl clusters, halophilic adaptation and catalysis of Haloferax mediterranei D-2-hydroxyacid dehydrogenase.
Domenech, J., Pramanpol, N., Bisson, C., Sedelnikova, S.E., Barrett, J.R., Dakhil, A.A.A.B., Mykhaylyk, V., Abdelhameed, A.S., Harding, S.E., Rice, D.W., Baker, P.J., Ferrer, J.(2025) Commun Biol 8: 1170-1170
- PubMed: 40770045
- DOI: https://doi.org/10.1038/s42003-025-08587-7
- Primary Citation of Related Structures:
5MH5, 5MH6, 5MHA, 8QZA, 8QZB, 9IBE - PubMed Abstract:
Enzymes from salt-in halophiles are stable in conditions of low water activity with applications in chiral synthesis requiring organic solvents, yet the origins of such stability remains poorly understood. Here we describe the molecular basis of the reaction mechanism and dual NADH/NADPH-specificity of D2HDH, a 2-hydroxyacid dehydrogenase from the extreme halophile Haloferax mediterranei, an organism whose proteins have to remain active in high intracellular concentrations of KCl. Halophilic adaptations of D2HDH include the expected acidic surface and a reduction in hydrophobic surface resulting from a lower lysine content. Structure determination of crystals of D2HDH grown with KCl showed that bound K + ions were coordinated predominantly by clusters of main chain protein carbonyl ligands, with no involvement of the numerous exposed surface carboxyls. Structural comparisons identified similar sites in other halophilic proteins suggesting that the generic use of carbonyl clusters to coordinate K + ions may also contribute in a carboxylate-independent way to the stabilisation of the folded state of the protein in its high salt environment.
- Dept. Bioquımica y Biología Molecular y EQA. Universidad de Alicante, Alicante, Spain.
Organizational Affiliation: