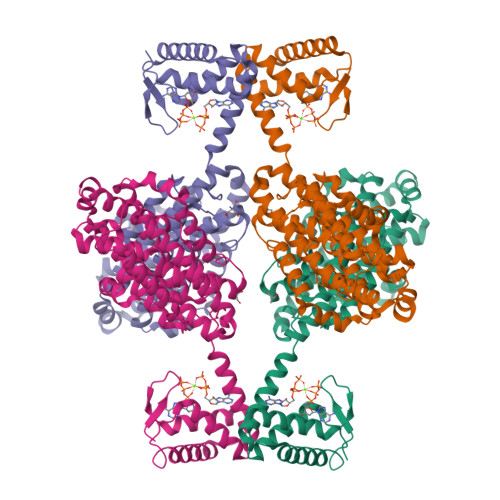
Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit.
Rozman Grinberg, I., Lundin, D., Hasan, M., Crona, M., Jonna, V.R., Loderer, C., Sahlin, M., Markova, N., Borovok, I., Berggren, G., Hofer, A., Logan, D.T., Sjoberg, B.M.(2018) Elife 7
- PubMed: 29388911
- DOI: https://doi.org/10.7554/eLife.31529
- Primary Citation of Related Structures:
5OLK - PubMed Abstract:
Ribonucleotide reductases (RNRs) are key enzymes in DNA metabolism, with allosteric mechanisms controlling substrate specificity and overall activity. In RNRs, the activity master-switch, the ATP-cone, has been found exclusively in the catalytic subunit. In two class I RNR subclasses whose catalytic subunit lacks the ATP-cone, we discovered ATP-cones in the radical-generating subunit. The ATP-cone in the Leeuwenhoekiella blandensis radical-generating subunit regulates activity via quaternary structure induced by binding of nucleotides. ATP induces enzymatically competent dimers, whereas dATP induces non-productive tetramers, resulting in different holoenzymes. The tetramer forms by interactions between ATP-cones, shown by a 2.45 Å crystal structure. We also present evidence for an Mn III Mn IV metal center. In summary, lack of an ATP-cone domain in the catalytic subunit was compensated by transfer of the domain to the radical-generating subunit. To our knowledge, this represents the first observation of transfer of an allosteric domain between components of the same enzyme complex.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden.