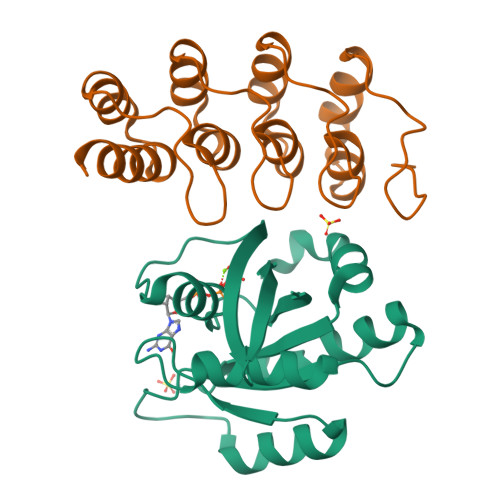
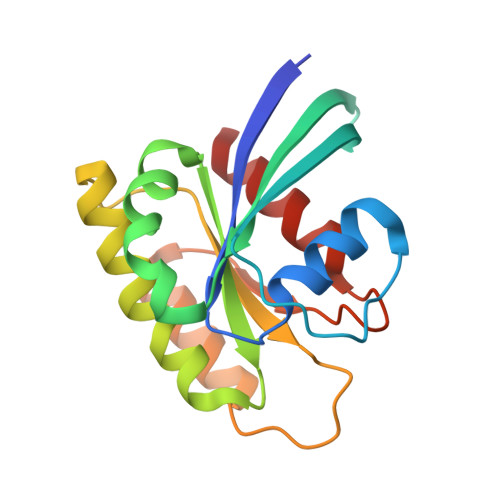
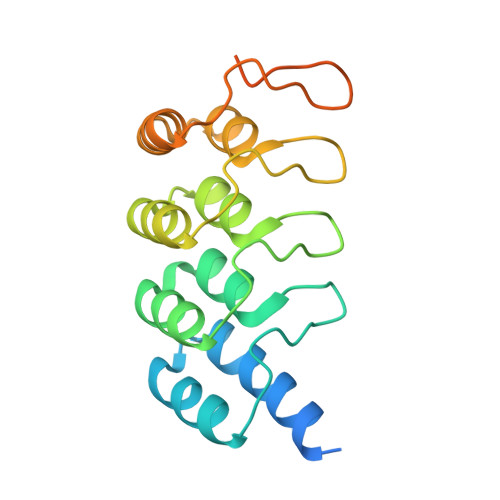
Inhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
Guillard, S., Kolasinska-Zwierz, P., Debreczeni, J., Breed, J., Zhang, J., Bery, N., Marwood, R., Tart, J., Overman, R., Stocki, P., Minstry, B., Phillips, C., Rabbitts, T., Jackson, R., Minter, R.To be published.