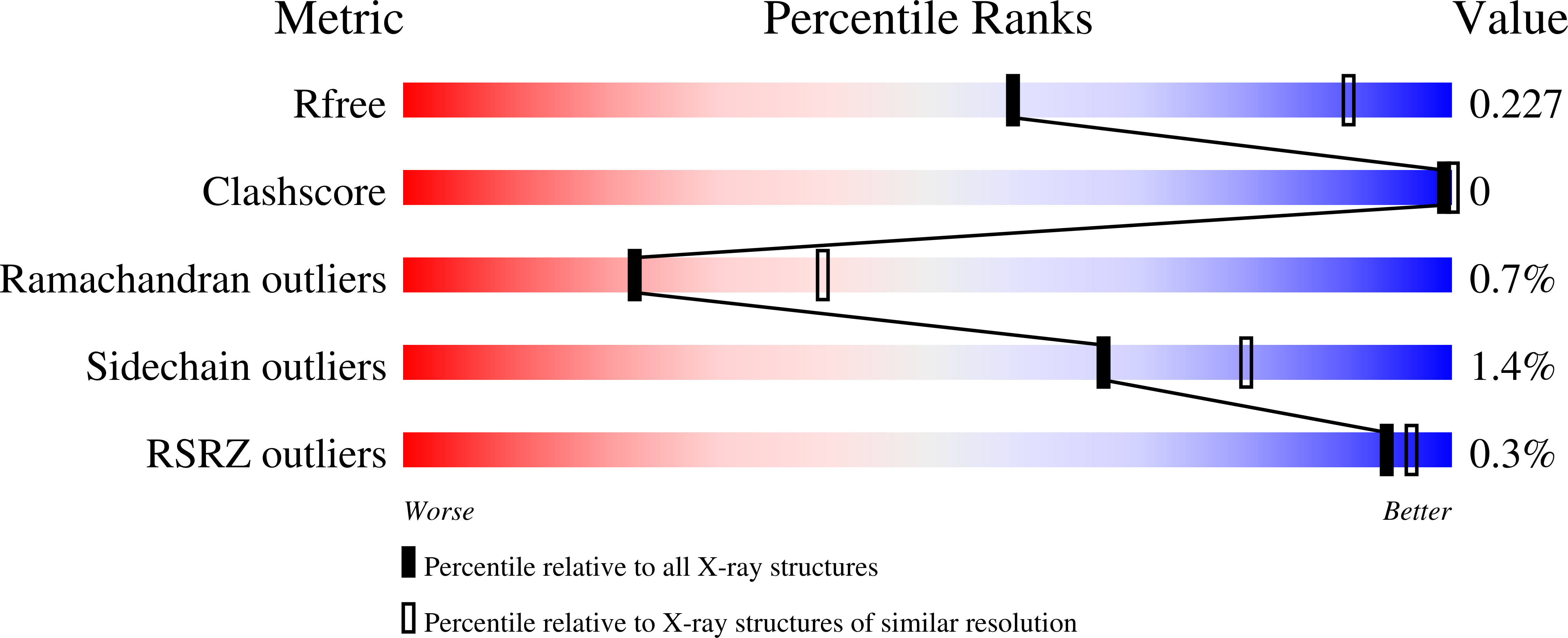
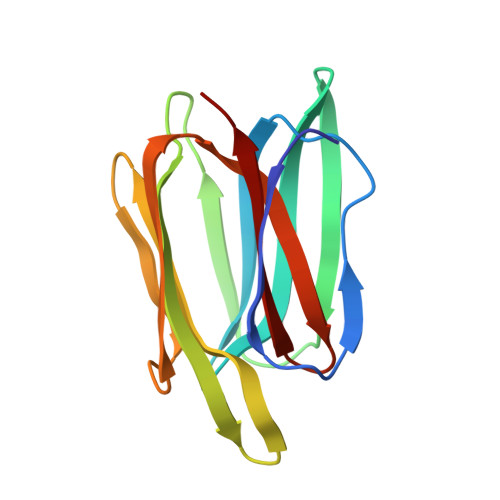
Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Azarkan, M., Feller, G., Vandenameele, J., Herman, R., El Mahyaoui, R., Sauvage, E., Vanden Broeck, A., Matagne, A., Charlier, P., Kerff, F.(2018) Sci Rep 8: 11508-11508
- PubMed: 30065388
- DOI: https://doi.org/10.1038/s41598-018-29439-x
- Primary Citation of Related Structures:
6FLW, 6FLY, 6FLZ - PubMed Abstract:
A mannose binding jacalin-related lectin from Ananas comosus stem (AcmJRL) was purified and biochemically characterized. This lectin is homogeneous according to native, SDS-PAGE and N-terminal sequencing and the theoretical molecular mass was confirmed by ESI-Q-TOF-MS. AcmJRL was found homodimeric in solution by size-exclusion chromatography. Rat erythrocytes are agglutinated by AcmJRL while no agglutination activity is detected against rabbit and sheep erythrocytes. Hemagglutination activity was found more strongly inhibited by mannooligomannosides than by D-mannose. The carbohydrate-binding specificity of AcmJRL was determined in some detail by isothermal titration calorimetry. All sugars tested were found to bind with low affinity to AcmJRL, with K a values in the mM range. In agreement with hemagglutination assays, the affinity increased from D-mannose to di-, tri- and penta-mannooligosaccharides. Moreover, the X-ray crystal structure of AcmJRL was obtained in an apo form as well as in complex with D-mannose and methyl-α-D-mannopyranoside, revealing two carbohydrate-binding sites per monomer similar to the banana lectin BanLec. The absence of a wall separating the two binding sites, the conformation of β7β8 loop and the hemagglutinating activity are reminiscent of the BanLec His84Thr mutant, which presents a strong anti-HIV activity in absence of mitogenic activity.
Organizational Affiliation:
Université Libre de Bruxelles, Faculty of Medicine, Protein Chemistry Unit, Campus Erasme (CP 609), 808 route de Lennik, 1070, Brussels, Belgium.