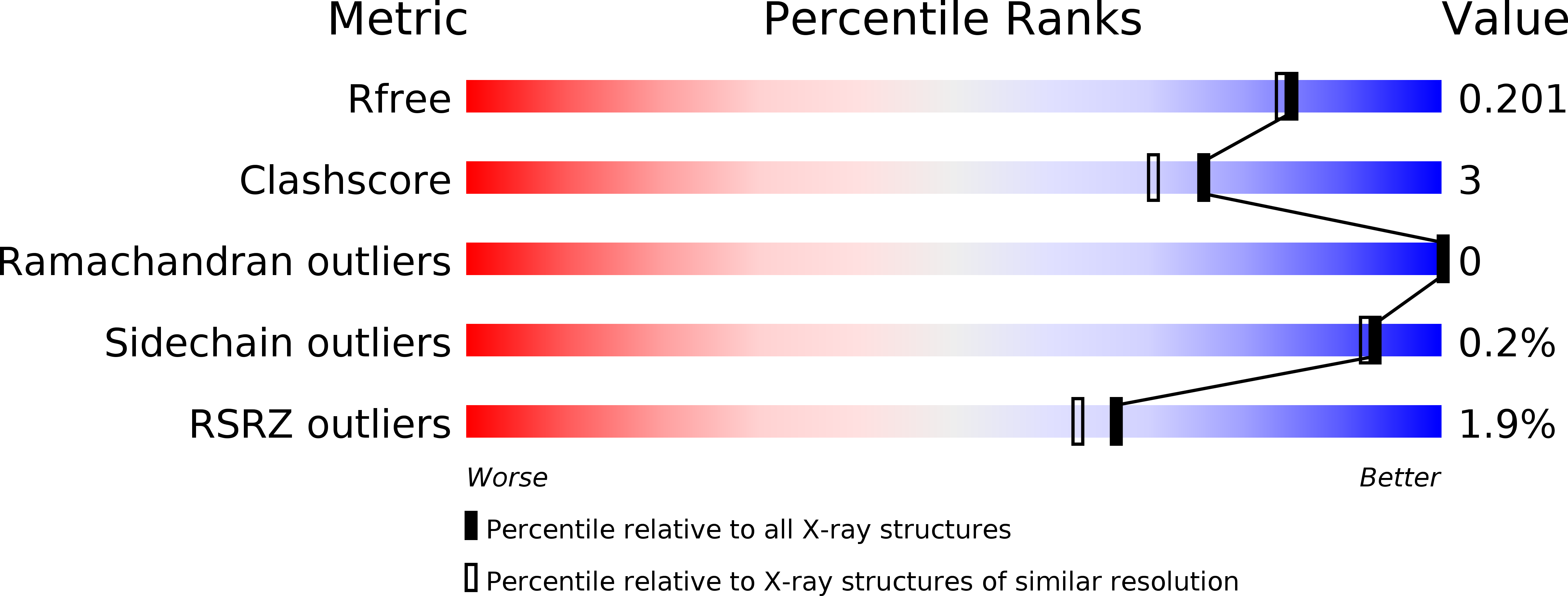
Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Smith, C.A., Stewart, N.K., Toth, M., Vakulenko, S.B.(2019) Antimicrob Agents Chemother 63
- PubMed: 31358584
- DOI: https://doi.org/10.1128/AAC.01202-19
- Primary Citation of Related Structures:
6P96, 6P97, 6P98, 6P99, 6P9C - PubMed Abstract:
Carbapenem-hydrolyzing class D carbapenemases (CHDLs) are enzymes that produce resistance to the last-resort carbapenem antibiotics, severely compromising the available therapeutic options for the treatment of life-threatening infections. A broad variety of CHDLs, including OXA-23, OXA-24/40, and OXA-58, circulate in Acinetobacter baumannii , while the OXA-48 CHDL is predominant in Enterobacteriaceae Extensive structural studies of A. baumannii enzymes have provided important information regarding their interactions with carbapenems and significantly contributed to the understanding of the mechanism of their carbapenemase activity. However, the interactions between carbapenems and OXA-48 have not yet been elucidated. We determined the X-ray crystal structures of the acyl-enzyme complexes of OXA-48 with four carbapenems, imipenem, meropenem, ertapenem, and doripenem, and compared them with those of known carbapenem complexes of A. baumannii CHDLs. In the A. baumannii enzymes, acylation by carbapenems triggers significant displacement of one of two conserved hydrophobic surface residues, resulting in the formation of a channel for entry of the deacylating water into the active site. We show that such a channel preexists in apo-OXA-48 and that only minor displacement of the conserved hydrophobic surface residues occurs upon the formation of OXA-48 acyl-enzyme intermediates. We also demonstrate that the extensive hydrophobic interactions that occur between a conserved hydrophobic bridge of the A. baumannii CHDLs and the carbapenem tails are lost in OXA-48 in the absence of an equivalent bridge structure. These data highlight significant differences between the interactions of carbapenems with OXA-48 and those with A. baumannii enzymes and provide important insights into the mechanism of carbapenemase activity of the major Enterobacteriaceae CHDL, OXA-48.
Organizational Affiliation:
Stanford Synchrotron Radiation Lightsource, Stanford University, Menlo Park, California, USA csmith@slac.stanford.edu svakulen@nd.edu.