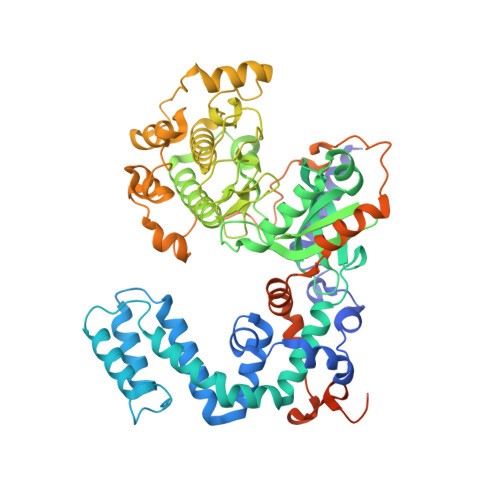
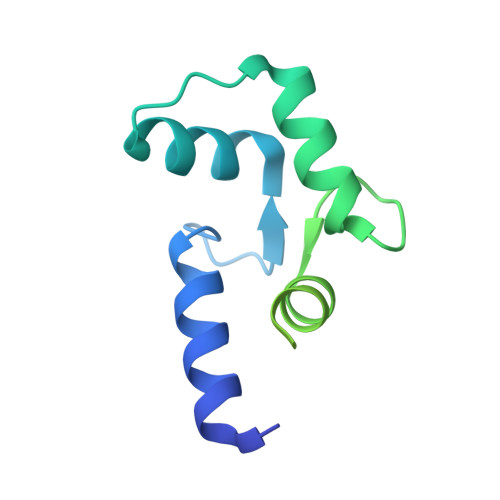
Structure of a GRK5-Calmodulin Complex Reveals Molecular Mechanism of GRK Activation and Substrate Targeting.
Komolov, K.E., Sulon, S.M., Bhardwaj, A., van Keulen, S.C., Duc, N.M., Laurinavichyute, D.K., Lou, H.J., Turk, B.E., Chung, K.Y., Dror, R.O., Benovic, J.L.(2021) Mol Cell 81: 323
- PubMed: 33321095
- DOI: https://doi.org/10.1016/j.molcel.2020.11.026
- Primary Citation of Related Structures:
6PJX - PubMed Abstract:
The phosphorylation of G protein-coupled receptors (GPCRs) by GPCR kinases (GRKs) facilitates arrestin binding and receptor desensitization. Although this process can be regulated by Ca 2+ -binding proteins such as calmodulin (CaM) and recoverin, the molecular mechanisms are poorly understood. Here, we report structural, computational, and biochemical analysis of a CaM complex with GRK5, revealing how CaM shapes GRK5 response to calcium. The CaM N and C domains bind independently to two helical regions at the GRK5 N and C termini to inhibit GPCR phosphorylation, though only the C domain interaction disrupts GRK5 membrane association, thereby facilitating cytoplasmic translocation. The CaM N domain strongly activates GRK5 via ordering of the amphipathic αN-helix of GRK5 and allosteric disruption of kinase-RH domain interaction for phosphorylation of cytoplasmic GRK5 substrates. These results provide a framework for understanding how two functional effects, GRK5 activation and localization, can cooperate under control of CaM for selective substrate targeting by GRK5.
- Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, PA 19107, USA.
Organizational Affiliation: