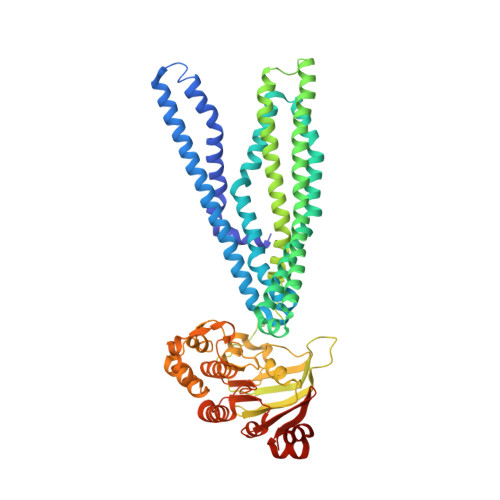
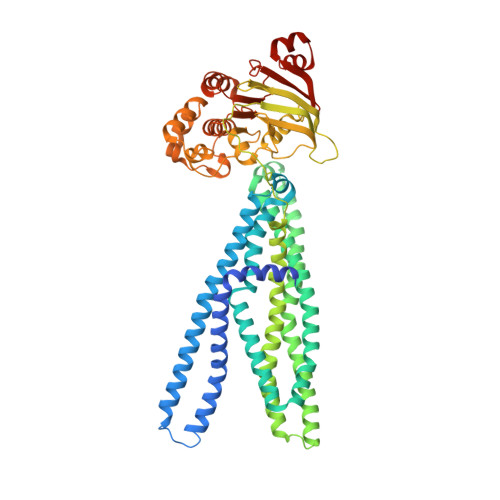
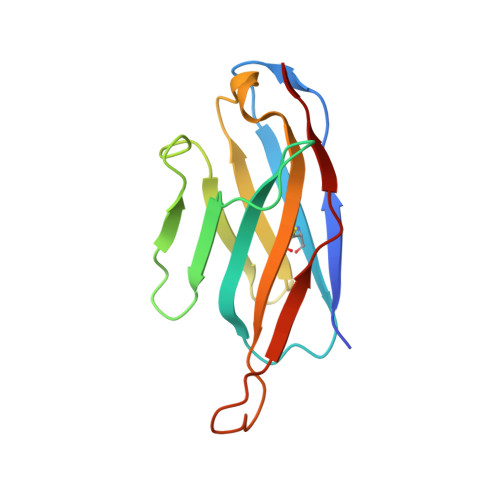
The extracellular gate shapes the energy profile of an ABC exporter.
Hutter, C.A.J., Timachi, M.H., Hurlimann, L.M., Zimmermann, I., Egloff, P., Goddeke, H., Kucher, S., Stefanic, S., Karttunen, M., Schafer, L.V., Bordignon, E., Seeger, M.A.(2019) Nat Commun 10: 2260-2260
- PubMed: 31113958
- DOI: https://doi.org/10.1038/s41467-019-09892-6
- Primary Citation of Related Structures:
6QUZ, 6QV0, 6QV1, 6QV2 - PubMed Abstract:
ABC exporters harness the energy of ATP to pump substrates across membranes. Extracellular gate opening and closure are key steps of the transport cycle, but the underlying mechanism is poorly understood. Here, we generated a synthetic single domain antibody (sybody) that recognizes the heterodimeric ABC exporter TM287/288 exclusively in the presence of ATP, which was essential to solve a 3.2 Å crystal structure of the outward-facing transporter. The sybody binds to an extracellular wing and strongly inhibits ATPase activity by shifting the transporter's conformational equilibrium towards the outward-facing state, as shown by double electron-electron resonance (DEER). Mutations that facilitate extracellular gate opening result in a comparable equilibrium shift and strongly reduce ATPase activity and drug transport. Using the sybody as conformational probe, we demonstrate that efficient extracellular gate closure is required to dissociate the NBD dimer after ATP hydrolysis to reset the transporter back to its inward-facing state.
- Institute of Medical Microbiology, University of Zurich, Gloriastr. 28/30, 8006, Zurich, Switzerland.
Organizational Affiliation: