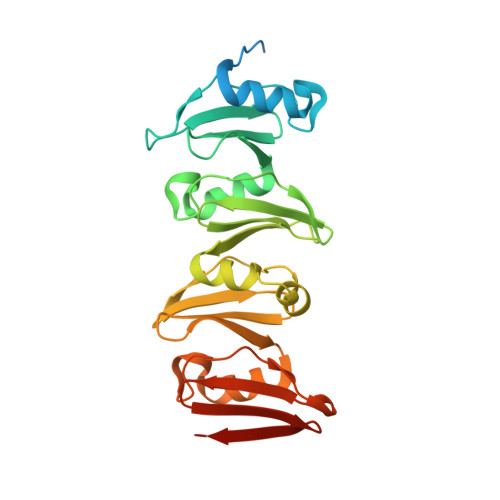
The C-terminal domain of Corynebacterium glutamicum mycoloyltransferase A is composed of five repeated motifs involved in cell wall binding and stability.
Dietrich, C., Li de la Sierra-Gallay, I., Masi, M., Girard, E., Dautin, N., Constantinesco-Becker, F., Tropis, M., Daffe, M., van Tilbeurgh, H., Bayan, N.(2020) Mol Microbiol 114: 1-16
- PubMed: 32073722
- DOI: https://doi.org/10.1111/mmi.14492
- Primary Citation of Related Structures:
6SWZ, 6SX4 - PubMed Abstract:
The genomes of Corynebacteriales contain several genes encoding mycoloyltransferases (Myt) that are specific cell envelope enzymes essential for the biogenesis of the outer membrane. MytA is a major mycoloyltransferase of Corynebacterium glutamicum, displaying an N-terminal domain with esterase activity and a C-terminal extension containing a conserved repeated Leu-Gly-Phe-Pro (LGFP) sequence motif of unknown function. This motif is highly conserved in Corynebacteriales and found associated with cell wall hydrolases and with proteins of unknown function. In this study, we determined the crystal structure of MytA and found that its C-terminal domain is composed of five LGFP motifs and forms a long stalk perpendicular to the N-terminal catalytic α/β-hydrolase domain. The LGFP motifs are composed of a 4-stranded β-fold and occupy alternating orientations along the axis of the stalk. Multiple acetate binding pockets were identified in the stalk, which could correspond to putative ligand-binding sites. By using various MytA mutants and complementary in vitro and in vivo approaches, we provide evidence that the C-terminal LGFP domain interacts with the cell wall peptidoglycan-arabinogalactan polymer. We also show that the C-terminal LGFP domain is not required for the activity of MytA but rather contributes to the overall integrity of the cell envelope.
- Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, Gif-sur-Yvette, France.
Organizational Affiliation: