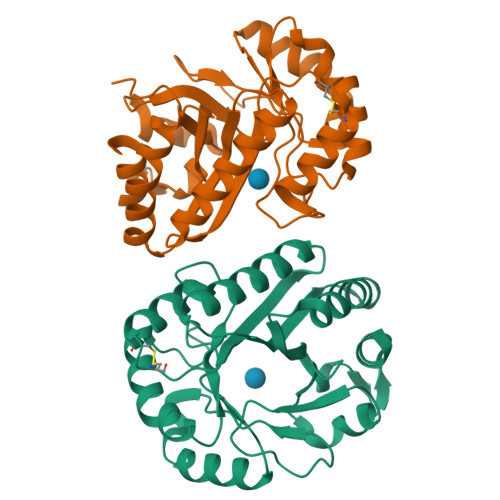
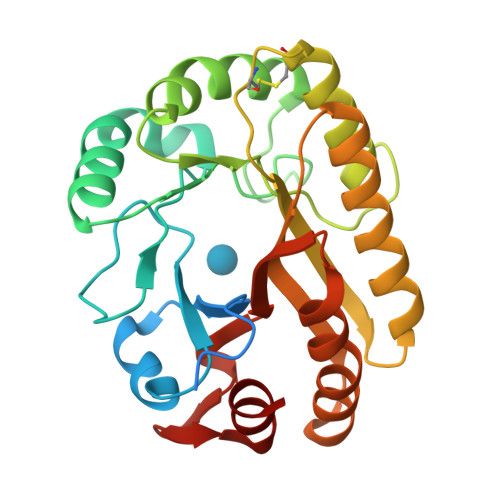
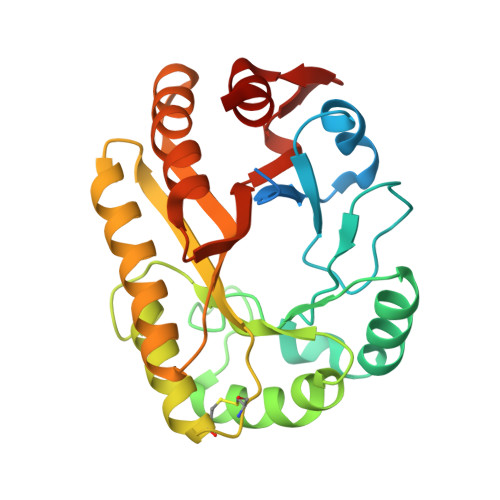
Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Santos, C.R., Costa, P.A.C.R., Vieira, P.S., Gonzalez, S.E.T., Correa, T.L.R., Lima, E.A., Mandelli, F., Pirolla, R.A.S., Domingues, M.N., Cabral, L., Martins, M.P., Cordeiro, R.L., Junior, A.T., Souza, B.P., Prates, E.T., Gozzo, F.C., Persinoti, G.F., Skaf, M.S., Murakami, M.T.(2020) Nat Chem Biol 16: 920-929
- PubMed: 32451508
- DOI: https://doi.org/10.1038/s41589-020-0554-5
- Primary Citation of Related Structures:
6UAQ, 6UAR, 6UAS, 6UAT, 6UAU, 6UAV, 6UAW, 6UAX, 6UAY, 6UAZ, 6UB0, 6UB1, 6UB2, 6UB3, 6UB4, 6UB5, 6UB6, 6UB7, 6UB8, 6UBA, 6UBB, 6UBC, 6UBD, 6UFL, 6UFZ - PubMed Abstract:
The fundamental and assorted roles of β-1,3-glucans in nature are underpinned on diverse chemistry and molecular structures, demanding sophisticated and intricate enzymatic systems for their processing. In this work, the selectivity and modes of action of a glycoside hydrolase family active on β-1,3-glucans were systematically investigated combining sequence similarity network, phylogeny, X-ray crystallography, enzyme kinetics, mutagenesis and molecular dynamics. This family exhibits a minimalist and versatile (α/β)-barrel scaffold, which can harbor distinguishing exo or endo modes of action, including an ancillary-binding site for the anchoring of triple-helical β-1,3-glucans. The substrate binding occurs via a hydrophobic knuckle complementary to the canonical curved conformation of β-1,3-glucans or through a substrate conformational change imposed by the active-site topology of some fungal enzymes. Together, these findings expand our understanding of the enzymatic arsenal of bacteria and fungi for the breakdown and modification of β-1,3-glucans, which can be exploited for biotechnological applications.
Organizational Affiliation:
Brazilian Biorenewables National Laboratory, Brazilian Center for Research in Energy and Materials, Campinas, São Paulo, Brazil.