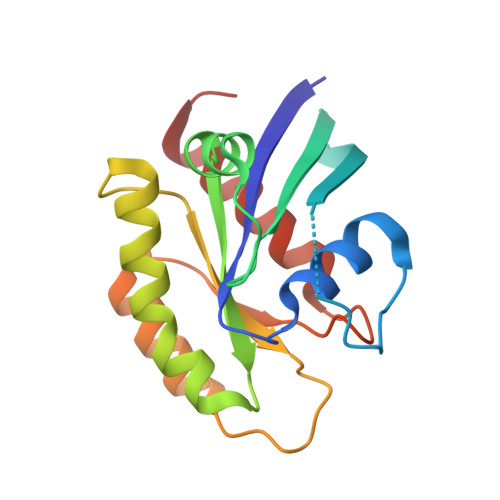
GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Zhang, Z., Gao, R., Hu, Q., Peacock, H., Peacock, D.M., Dai, S., Shokat, K.M., Suga, H.(2020) ACS Cent Sci 6: 1753-1761
- PubMed: 33145412
- DOI: https://doi.org/10.1021/acscentsci.0c00514
- Primary Citation of Related Structures:
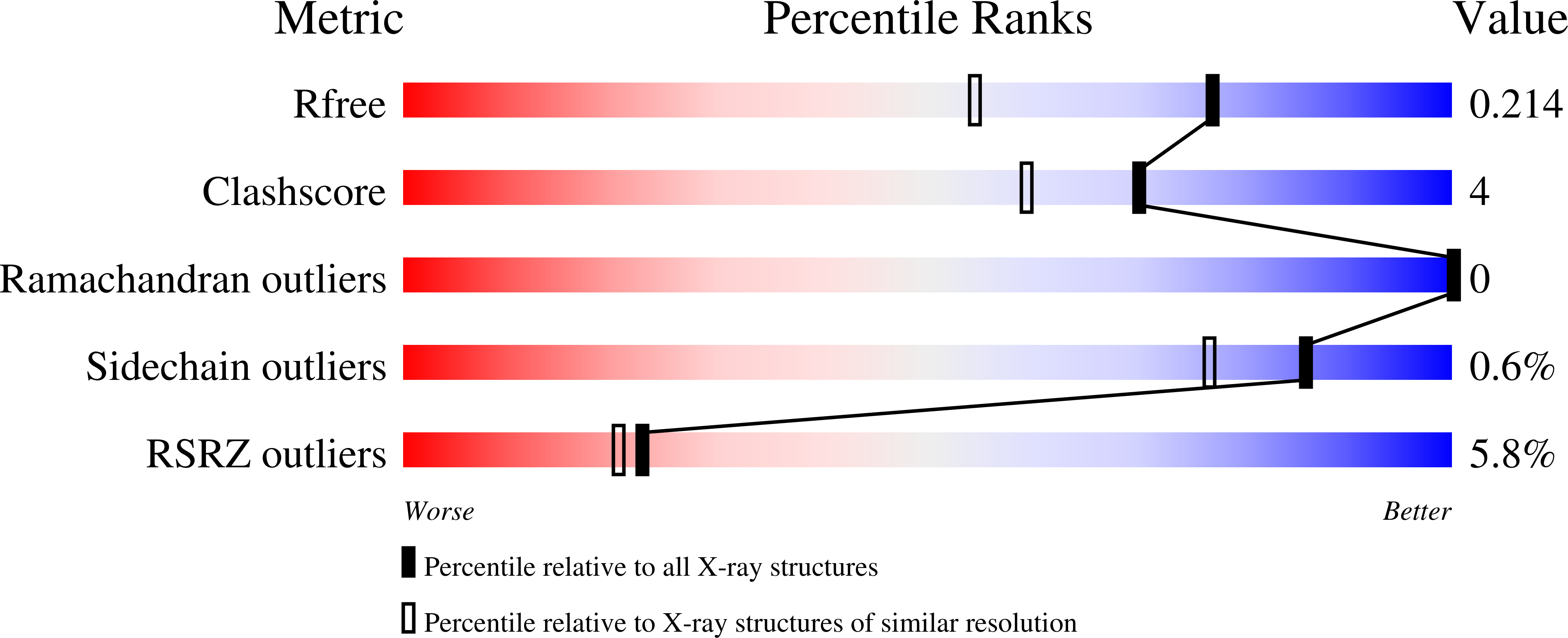
6WGN - PubMed Abstract:
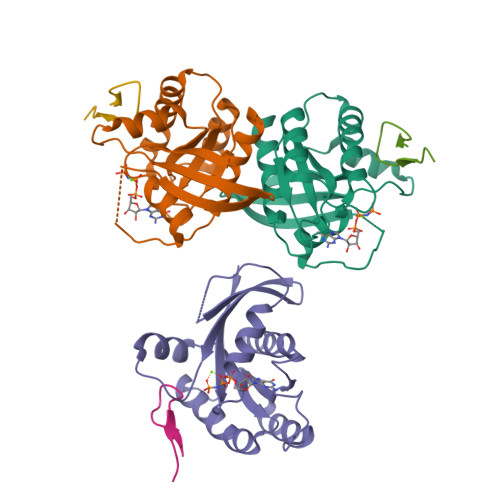
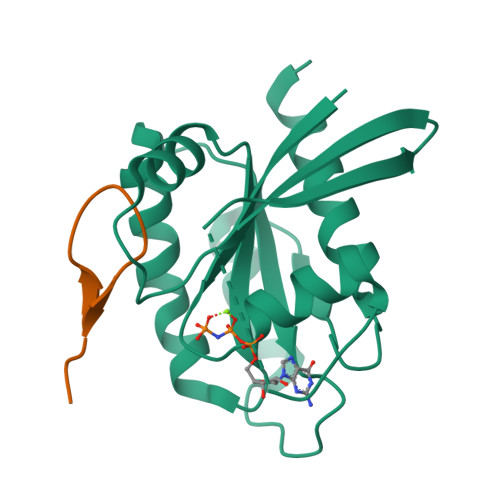
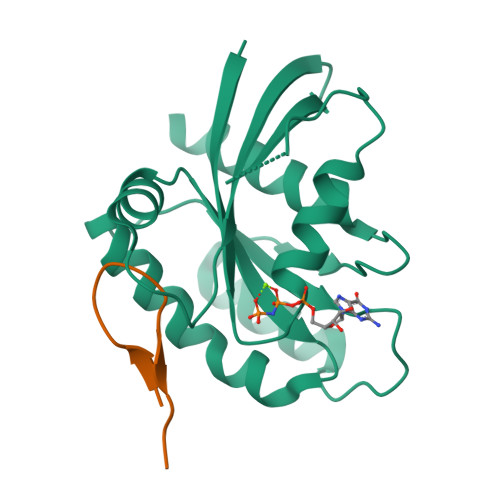
We report the identification of three cyclic peptide ligands of K-Ras(G12D) using an integrated in vitro translation-mRNA display selection platform. These cyclic peptides show preferential binding to the GTP-bound state of K-Ras(G12D) over the GDP-bound state and block Ras-Raf interaction. A co-crystal structure of peptide KD2 with K-Ras(G12D)·GppNHp reveals that this peptide binds in the Switch II groove region with concomitant opening of the Switch II loop and a 40° rotation of the α2 helix, and that a threonine residue (Thr10) on KD2 has direct access to the mutant aspartate (Asp12) on K-Ras. Replacing this threonine with non-natural amino acids afforded peptides with improved potency at inhibiting the interaction between Raf1-RBD and K-Ras(G12D) but not wildtype K-Ras. The union of G12D over wildtype selectivity and GTP state/GDP state selectivity is particularly desirable, considering that oncogenic K-Ras(G12D) exists predominantly in the GTP state in cancer cells, and wildtype K-Ras signaling is important for the maintenance of healthy cells.
Organizational Affiliation:
Department of Cellular and Molecular Pharmacology, Howard Hughes Medical Institute, University of California-San Francisco, San Francisco, California 94158, United States.