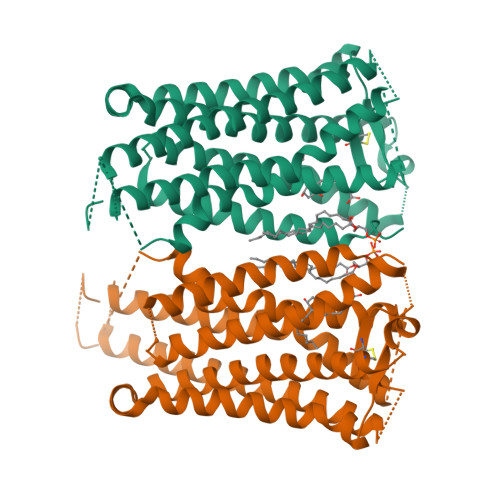
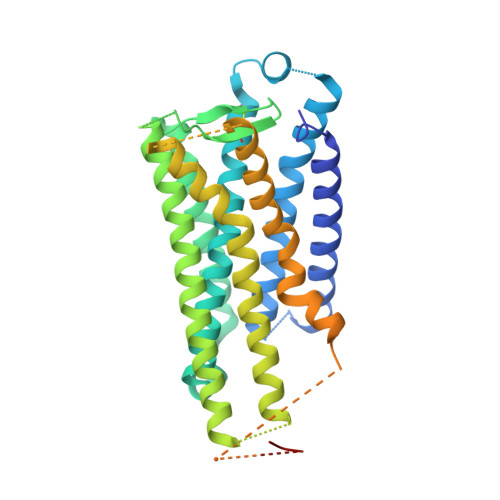
Crystal structure of the endogenous agonist-bound prostanoid receptor EP3.
Morimoto, K., Suno, R., Hotta, Y., Yamashita, K., Hirata, K., Yamamoto, M., Narumiya, S., Iwata, S., Kobayashi, T.(2019) Nat Chem Biol 15: 8-10
- PubMed: 30510192
- DOI: https://doi.org/10.1038/s41589-018-0171-8
- Primary Citation of Related Structures:
6AK3 - PubMed Abstract:
Prostanoids are a series of bioactive lipid metabolites that function in an autacoid manner via activation of cognate G-protein-coupled receptors (GPCRs). Here, we report the crystal structure of human prostaglandin (PG) E receptor subtype EP3 bound to endogenous ligand PGE 2 at 2.90 Å resolution. The structure reveals important insights into the activation mechanism of prostanoid receptors and provides a molecular basis for the binding modes of endogenous ligands.
Organizational Affiliation:
Department of Cell Biology, Graduate School of Medicine, Kyoto University, Konoe-cho, Yoshida, Sakyo-ku, Kyoto, Japan. k.morimoto@mfour.med.kyoto-u.ac.jp.