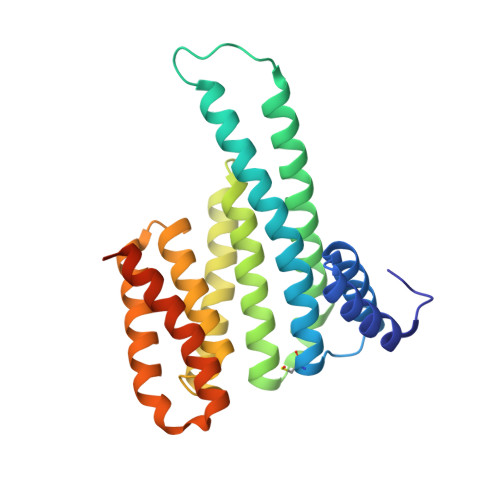
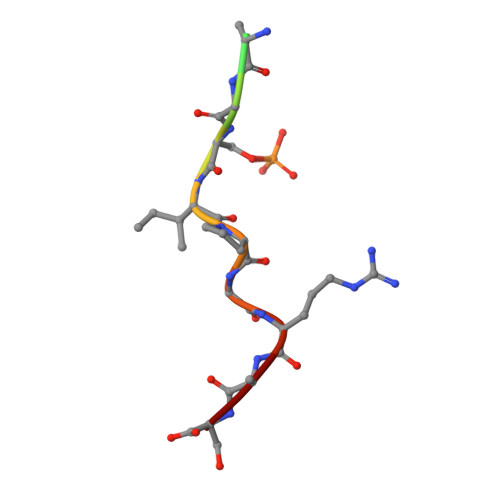
An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
Wolter, M., Valenti, D., Cossar, P.J., Hristeva, S., Levy, L.M., Genski, T., Hoffmann, T., Brunsveld, L., Tzalis, D., Ottmann, C.(2021) J Med Chem 64: 8423-8436
- PubMed: 34076416
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00401
- Primary Citation of Related Structures:
7BI3, 7BIQ, 7BIW, 7BIY, 7BJB, 7BJF, 7BJL, 7BJW, 7BKH, 7NJ9, 7NJB, 7NK3, 7NK5, 7NLA, 7NLE, 7NM1, 7NM3, 7NM9, 7NMH, 7NR7, 7NV4, 7NVI, 7NWS, 7NXS, 7NXT, 7NXW, 7NXY, 7NY4, 7NYE, 7NYF, 7NYG, 7NZ6, 7NZG, 7NZK, 7NZV, 7O34, 7O3A, 7O3F, 7O3P, 7O3Q, 7O3R, 7O3S, 7O57, 7O59, 7O5A, 7O5C, 7O5D, 7O5F, 7O5G, 7O5O - PubMed Abstract:
Protein-protein modulation has emerged as a proven approach to drug discovery. While significant progress has been gained in developing protein-protein interaction (PPI) inhibitors, the orthogonal approach of PPI stabilization lacks established methodologies for drug design. Here, we report the systematic ″bottom-up″ development of a reversible covalent PPI stabilizer. An imine bond was employed to anchor the stabilizer at the interface of the 14-3-3/p65 complex, leading to a molecular glue that elicited an 81-fold increase in complex stabilization. Utilizing protein crystallography and biophysical assays, we deconvoluted how chemical properties of a stabilizer translate to structural changes in the ternary 14-3-3/p65/molecular glue complex. Furthermore, we explore how this leads to high cooperativity and increased stability of the complex.
- Department of Biomedical Engineering, Laboratory of Chemical Biology and Institute for Complex Molecular Systems, Eindhoven University of Technology, P.O. Box 513, 5600 MB Eindhoven, The Netherlands.
Organizational Affiliation: