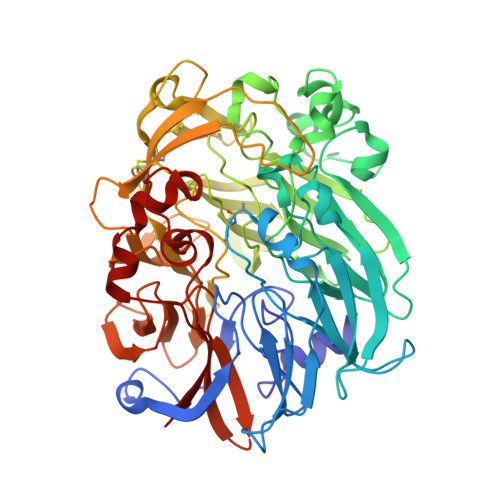
Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
Chan, S.I., Chuankhayan, P., Reddy Nareddy, P.K., Tsai, I.K., Tsai, Y.F., Chen, K.H., Yu, S.S., Chen, C.J.(2021) J Am Chem Soc 143: 3359-3372
- PubMed: 33629832
- DOI: https://doi.org/10.1021/jacs.0c11414
- Primary Citation of Related Structures:
7CDL, 7CE5, 7CE9, 7CED, 7CFX - PubMed Abstract:
The active site of methanol dehydrogenase (MDH) contains a rare disulfide bridge between adjacent cysteine residues. As a vicinal disulfide, the structure is highly strained, suggesting it might work together with the pyrroloquinoline quinone (PQQ) prosthetic group and the Ca 2+ ion in the catalytic turnover during methanol (CH 3 OH) oxidation. We purify MDH from Methylococcus capsulatus (Bath) with the disulfide bridge broken into two thiols. Spectroscopic and high-resolution X-ray crystallographic studies of this form of MDH indicate that the disulfide bridge is redox active. We observe an internal redox process within the holo -MDH that produces a disulfide radical anion concomitant with a companion PQQ radical, as evidenced by an optical absorption at 408 nm and a magnetically dipolar-coupled biradical in the EPR spectrum. These observations are corroborated by electron-density changes between the two cysteine sulfurs of the disulfide bridge as well as between the bound Ca 2+ ion and the O5-C5 bond of the PQQ in the high-resolution X-ray structure. On the basis of these findings, we propose a mechanism for the controlled redistribution of the two electrons during hydride transfer from the CH 3 OH in the alcohol oxidation without formation of the reduced PQQ ethenediol, a biradical mechanism that allows for possible recovery of the hydride for transfer to an external NAD + oxidant in the regeneration of the PQQ cofactor for multiple catalytic turnovers. In support of this mechanism, a steady-state level of the disulfide radical anion is observed during turnover of the MDH in the presence of CH 3 OH and NAD + .
- Institute of Chemistry, Academia Sinica, Nangang, Taipei 11529, Taiwan.
Organizational Affiliation: