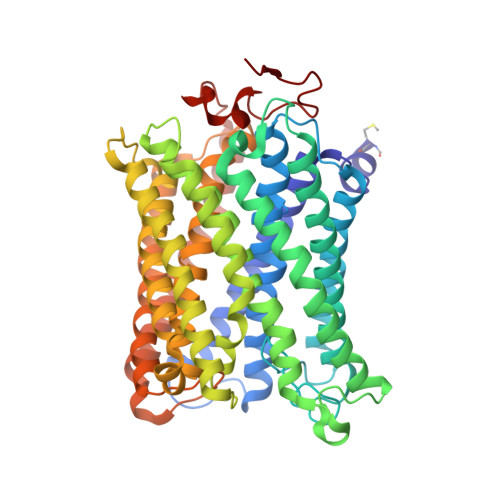
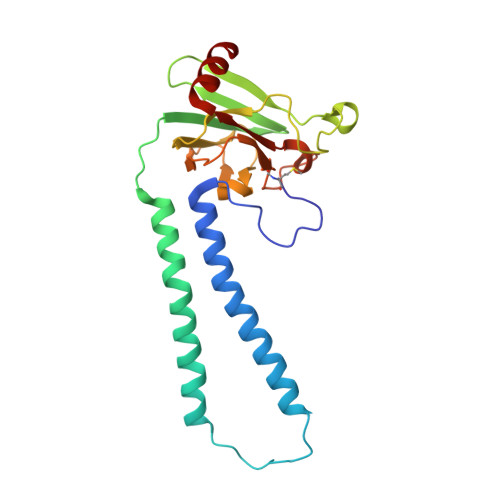
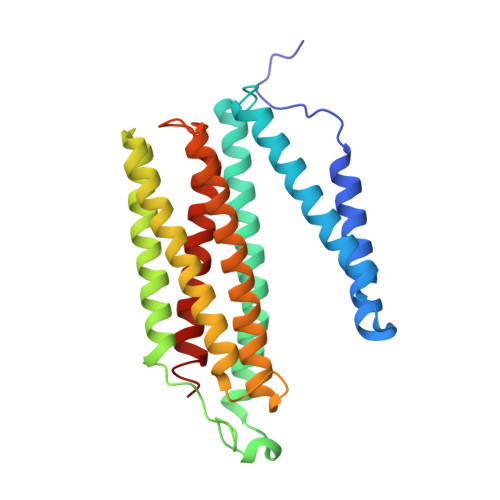
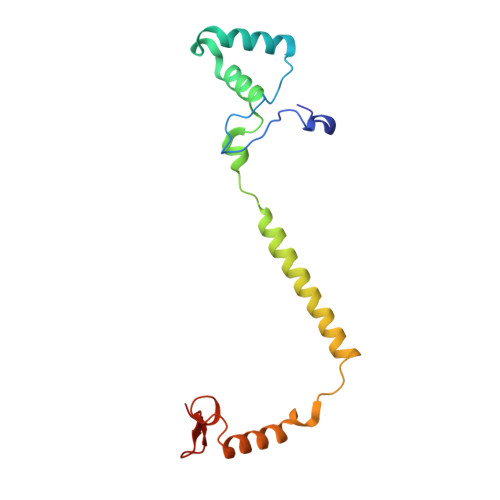
Critical roles of the Cu B site in efficient proton pumping as revealed by crystal structures of mammalian cytochrome c oxidase catalytic intermediates.
Shimada, A., Hara, F., Shinzawa-Itoh, K., Kanehisa, N., Yamashita, E., Muramoto, K., Tsukihara, T., Yoshikawa, S.(2021) J Biological Chem 297: 100967-100967
- PubMed: 34274318
- DOI: https://doi.org/10.1016/j.jbc.2021.100967
- Primary Citation of Related Structures:
7CP5, 7D5W, 7D5X - PubMed Abstract:
Mammalian cytochrome c oxidase (CcO) reduces O 2 to water in a bimetallic site including Fe a3 and Cu B giving intermediate molecules, termed A-, P-, F-, O-, E-, and R-forms. From the P-form on, each reaction step is driven by single-electron donations from cytochrome c coupled with the pumping of a single proton through the H-pathway, a proton-conducting pathway composed of a hydrogen-bond network and a water channel. The proton-gradient formed is utilized for ATP production by F-ATPase. For elucidation of the proton pumping mechanism, crystal structural determination of these intermediate forms is necessary. Here we report X-ray crystallographic analysis at ∼1.8 Å resolution of fully reduced CcO crystals treated with O 2 for three different time periods. Our disentanglement of intermediate forms from crystals that were composed of multiple forms determined that these three crystallographic data sets contained ∼45% of the O-form structure, ∼45% of the E-form structure, and ∼20% of an oxymyoglobin-type structure consistent with the A-form, respectively. The O- and E-forms exhibit an unusually long Cu B 2+ -OH - distance and Cu B 1+ -H 2 O structure keeping Fe a3 3+ -OH - state, respectively, suggesting that the O- and E-forms have high electron affinities that cause the O→E and E→R transitions to be essentially irreversible and thus enable tightly coupled proton pumping. The water channel of the H-pathway is closed in the O- and E-forms and partially open in the R-form. These structures, together with those of the recently reported P- and F-forms, indicate that closure of the H-pathway water channel avoids back-leaking of protons for facilitating the effective proton pumping.
- Picobiology Institute, Graduate School of Life Science, University of Hyogo, kamigori, Akoh, Hyogo, Japan.
Organizational Affiliation: