Structural basis for sterol sensing by Scap and Insig
Yan, R., Cao, P., Song, W., Li, Y., Wang, T., Qian, H., Yan, C., Yan, N.(2021) Cell Rep 35: 109299
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Insulin-induced gene 2 protein | 225 | Homo sapiens | Mutation(s): 3 Gene Names: INSIG2 Membrane Entity: Yes | 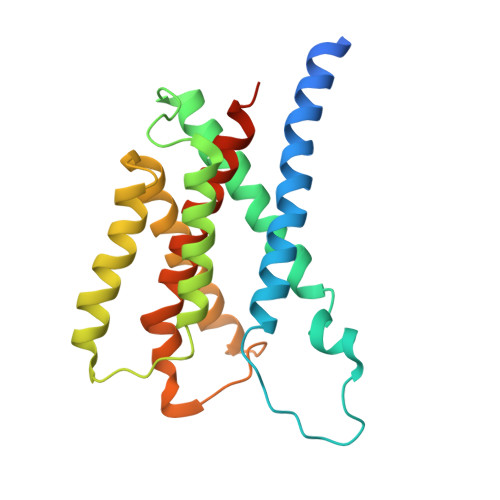 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y5U4 (Homo sapiens) Explore Q9Y5U4 Go to UniProtKB: Q9Y5U4 | |||||
PHAROS: Q9Y5U4 GTEx: ENSG00000125629 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y5U4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sterol regulatory element-binding protein cleavage-activating protein | 735 | Homo sapiens | Mutation(s): 0 Gene Names: SCAP, KIAA0199, PSEC0227 Membrane Entity: Yes | 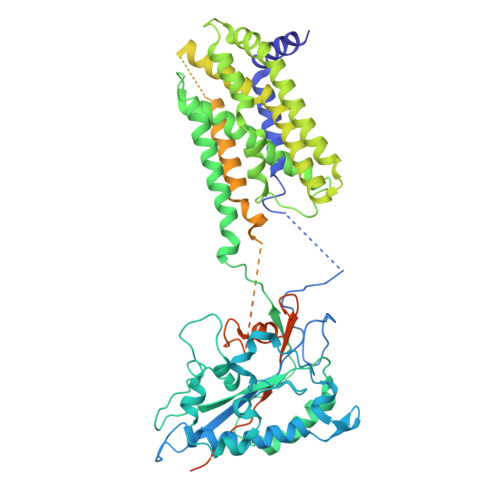 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q12770 (Homo sapiens) Explore Q12770 Go to UniProtKB: Q12770 | |||||
PHAROS: Q12770 GTEx: ENSG00000114650 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12770 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: Q12770-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AJP (Subject of Investigation/LOI) Query on AJP | D [auth A], E [auth A], F [auth A], G [auth A] | Digitonin C56 H92 O29 UVYVLBIGDKGWPX-KUAJCENISA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 3.0 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Science and Technology (MoST, China) | China | -- |