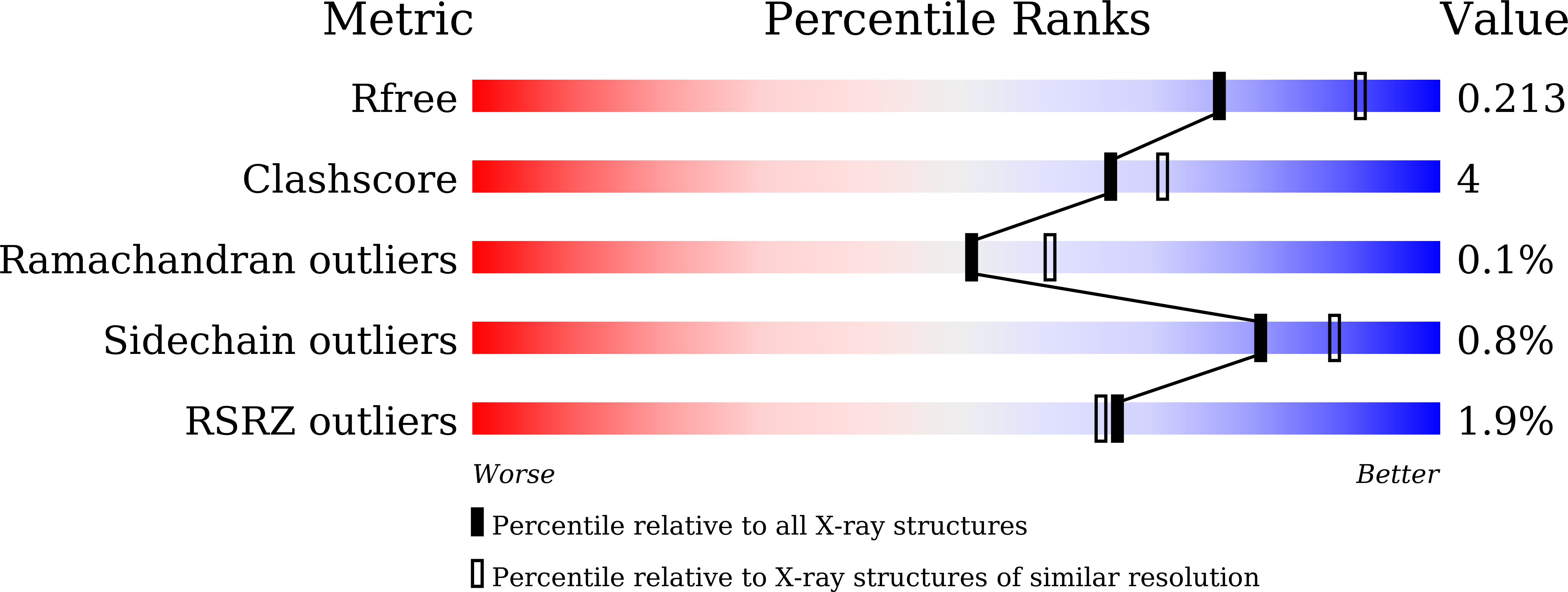
Crystal Structure and Functional Characterization of the Bifunctional N -(5'-Phosphoribosyl)anthranilate Isomerase-indole-3-glycerol-phosphate Synthase from Corynebacterium glutamicum
Park, W., Son, H.F., Lee, D., Kim, I.K., Kim, K.J.(2021) J Agric Food Chem 69: 12485-12493
- PubMed: 34657425
- DOI: https://doi.org/10.1021/acs.jafc.1c05132
- Primary Citation of Related Structures:
7ETX, 7ETY - PubMed Abstract:
L-Tryptophan is known as an aromatic amino acid and one of the essential amino acids that must be ingested through various additives or food. TrpCF is a bifunctional enzyme that has indole-glycerol-phosphate synthase (IGPS) and phosphoribosylanthranilate isomerase (PRAI) activity. In this report, we identified the crystal structure of TrpCF from Corynebacterium glutamicum ( Cg TrpCF) and successfully elucidated the active site by attaching rCdRP similar to the substrate and product of the TrpCF reaction. Also, we revealed that Cg TrpCF shows a conformational change at the loops upon substrate binding. We analyzed amino acid sequences of the homologues of Cg TrpCF, and the residues of the substrate-binding site in TrpCF were highly conserved except for some residues. These less conserved residues were replaced by site-directed mutagenesis experiments. Consequently, we obtained the Cg TrpCF P294K (PRAI CD/P294K ) variant that has enhanced activity.
Organizational Affiliation:
School of Life Sciences, BK21 Four KNU Creative BioResearch Group, KNU Institute for Microorganisms, Kyungpook National University, Daegu 41566, Republic of Korea.