7JHG
Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody
- PDB DOI: https://doi.org/10.2210/pdb7JHG/pdb
- EM Map EMD-22336: EMDB EMDataResource
- Classification: TRANSFERASE/IMMUNE SYSTEM
- Organism(s): Homo sapiens, Escherichia coli K-12, synthetic construct
- Expression System: Escherichia coli K-12
- Mutation(s): No
- Deposited: 2020-07-20 Released: 2021-07-21
- Funding Organization(s): National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS)
Experimental Data Snapshot
- Method: ELECTRON MICROSCOPY
- Resolution: 3.47 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
wwPDB Validation 3D Report Full Report
This is version 1.1 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'-AMP-activated protein kinase catalytic subunit alpha-1 | 484 | Homo sapiens | Mutation(s): 0 Gene Names: PRKAA1, AMPK1 EC: 2.7.11.1 (PDB Primary Data), 2.7.11.27 (PDB Primary Data), 2.7.11.31 (PDB Primary Data), 2.7.11.26 (PDB Primary Data) | 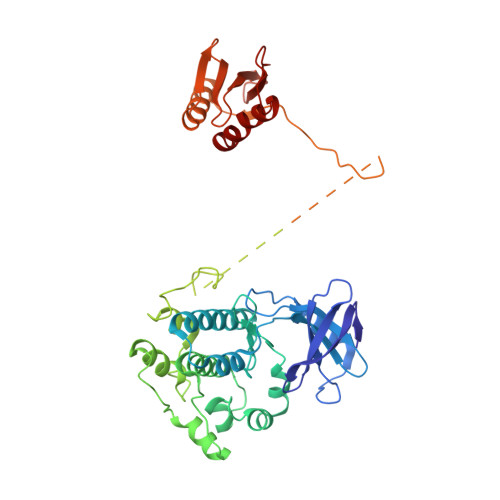 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13131 (Homo sapiens) Explore Q13131 Go to UniProtKB: Q13131 | |||||
PHAROS: Q13131 GTEx: ENSG00000132356 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13131 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'-AMP-activated protein kinase subunit beta-2 | 198 | Homo sapiens | Mutation(s): 0 Gene Names: PRKAB2 | 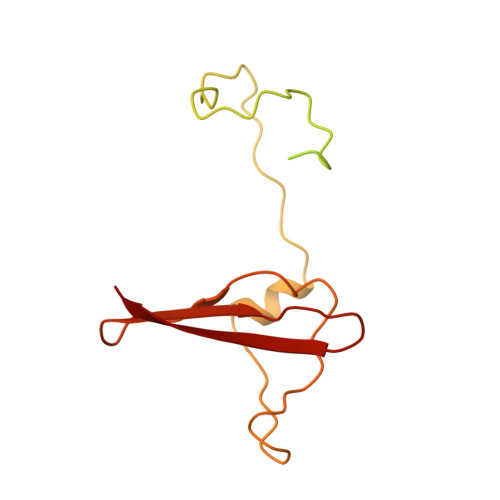 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O43741 (Homo sapiens) Explore O43741 Go to UniProtKB: O43741 | |||||
PHAROS: O43741 GTEx: ENSG00000131791 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O43741 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'-AMP-activated protein kinase subunit gamma-1 | C [auth G] | 306 | Homo sapiens | Mutation(s): 0 Gene Names: PRKAG1 | 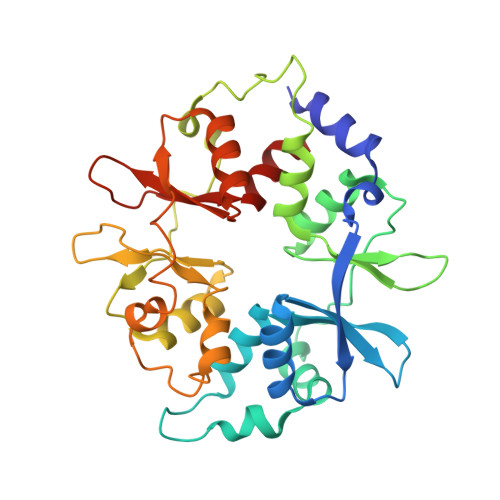 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P54619 (Homo sapiens) Explore P54619 Go to UniProtKB: P54619 | |||||
PHAROS: P54619 GTEx: ENSG00000181929 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P54619 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Maltodextrin-binding protein | D [auth M] | 373 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: malE | 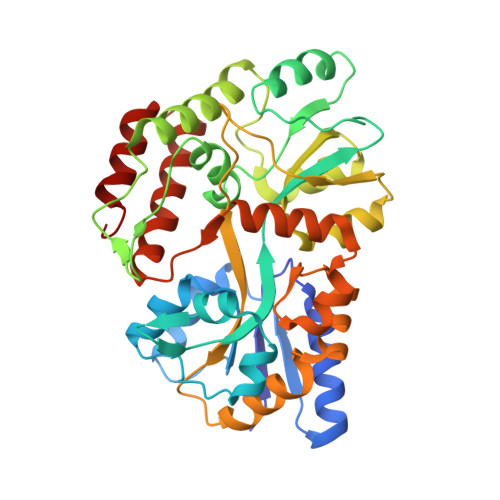 |
UniProt | |||||
Find proteins for P0AEX9 (Escherichia coli (strain K12)) Explore P0AEX9 Go to UniProtKB: P0AEX9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AEX9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fab light chain | E [auth L] | 215 | synthetic construct | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fab heavy chain | F [auth H] | 237 | synthetic construct | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody | G [auth N] | 159 | synthetic construct | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Oligosaccharides
Small Molecules
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | J [auth G] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| ADP Query on ADP | K [auth G] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| TAK (Subject of Investigation/LOI) Query on TAK | I [auth A] | 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine C24 H25 N5 O XHBVYDAKJHETMP-UHFFFAOYSA-N |  | ||
| AMP Query on AMP | L [auth G] | ADENOSINE MONOPHOSPHATE C10 H14 N5 O7 P UDMBCSSLTHHNCD-KQYNXXCUSA-N |  | ||
Biologically Interesting Molecules (External Reference) 1 Unique
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900001 Query on PRD_900001 | H [auth D] | alpha-maltose | Oligosaccharide / Nutrient |  | |
Experimental Data & Validation
Experimental Data
- Method: ELECTRON MICROSCOPY
- Resolution: 3.47 Å
- Aggregation State: PARTICLE
- Reconstruction Method: SINGLE PARTICLE
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX |
Entry History & Funding Information
Deposition Data
- Released Date: 2021-07-21 Deposition Author(s): Yan, Y., Murkherjee, S., Zhou, X.E., Xu, T.H., Xu, H.E., Kossiakoff, A.A., Melcher, K.
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01 GM117372 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01 GM129436 |
Revision History (Full details and data files)
- Version 1.0: 2021-07-21
Type: Initial release - Version 1.1: 2021-12-15
Changes: Database references














