Predicting the structural basis of targeted protein degradation by integrating molecular dynamics simulations with structural mass spectrometry.
Dixon, T., MacPherson, D., Mostofian, B., Dauzhenka, T., Lotz, S., McGee, D., Shechter, S., Shrestha, U.R., Wiewiora, R., McDargh, Z.A., Pei, F., Pal, R., Ribeiro, J.V., Wilkerson, T., Sachdeva, V., Gao, N., Jain, S., Sparks, S., Li, Y., Vinitsky, A., Zhang, X., Razavi, A.M., Kolossvary, I., Imbriglio, J., Evdokimov, A., Bergeron, L., Zhou, W., Adhikari, J., Ruprecht, B., Dickson, A., Xu, H., Sherman, W., Izaguirre, J.A.(2022) Nat Commun 13: 5884-5884
- PubMed: 36202813
- DOI: https://doi.org/10.1038/s41467-022-33575-4
- Primary Citation of Related Structures:
7S4E - PubMed Abstract:
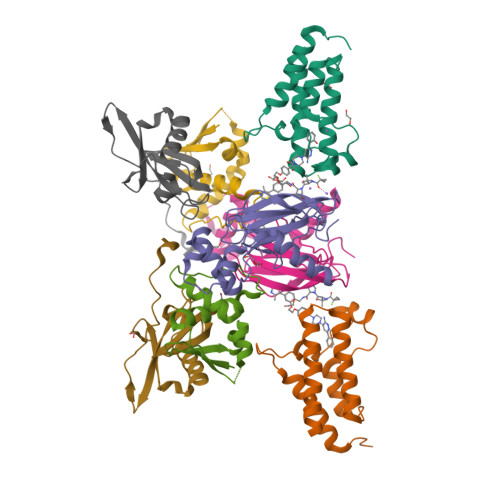
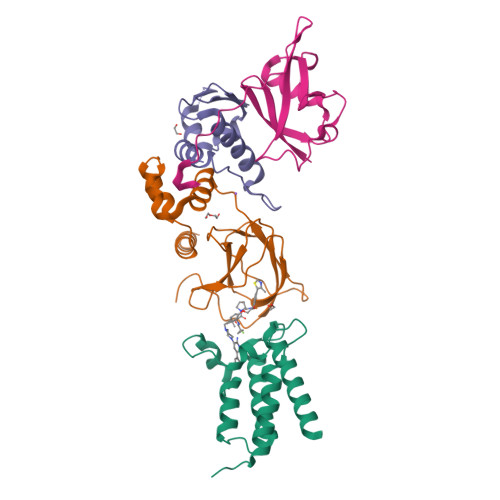
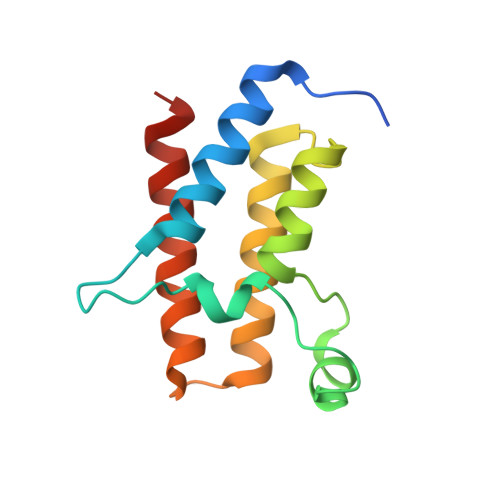
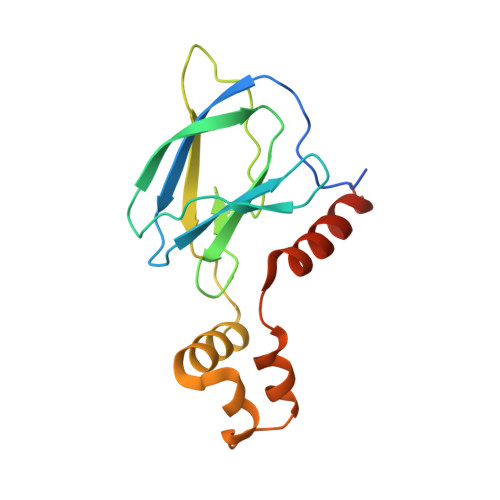
Targeted protein degradation (TPD) is a promising approach in drug discovery for degrading proteins implicated in diseases. A key step in this process is the formation of a ternary complex where a heterobifunctional molecule induces proximity of an E3 ligase to a protein of interest (POI), thus facilitating ubiquitin transfer to the POI. In this work, we characterize 3 steps in the TPD process. (1) We simulate the ternary complex formation of SMARCA2 bromodomain and VHL E3 ligase by combining hydrogen-deuterium exchange mass spectrometry with weighted ensemble molecular dynamics (MD). (2) We characterize the conformational heterogeneity of the ternary complex using Hamiltonian replica exchange simulations and small-angle X-ray scattering. (3) We assess the ubiquitination of the POI in the context of the full Cullin-RING Ligase, confirming experimental ubiquitinomics results. Differences in degradation efficiency can be explained by the proximity of lysine residues on the POI relative to ubiquitin.
Organizational Affiliation:
Roivant Discovery, New York City, NY, 10036, USA.