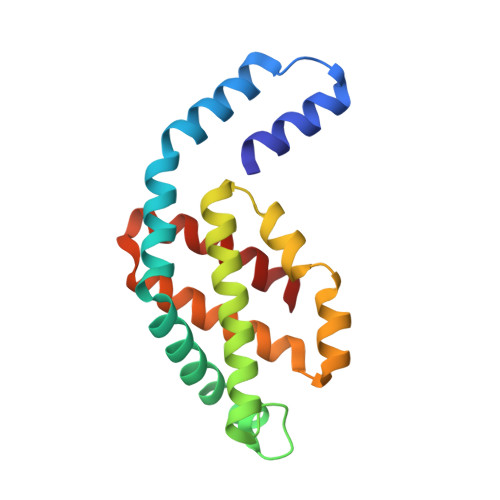
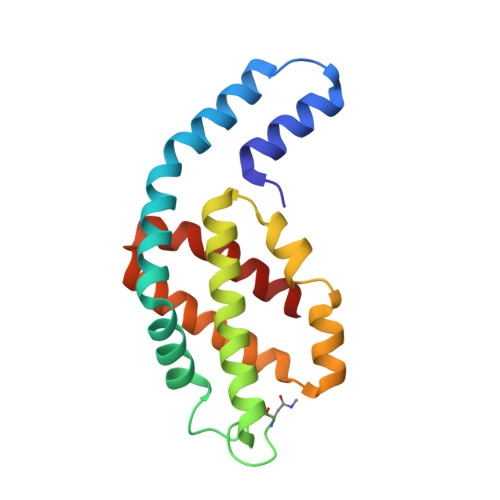
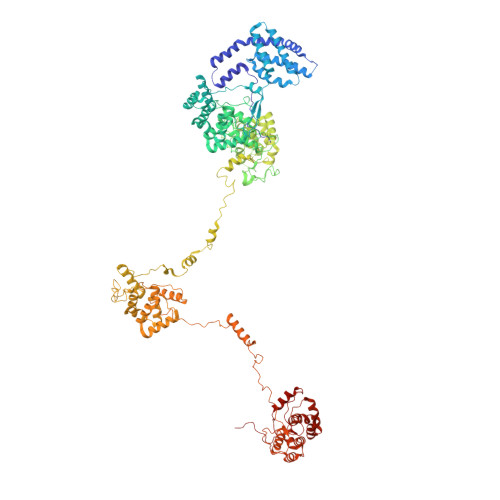
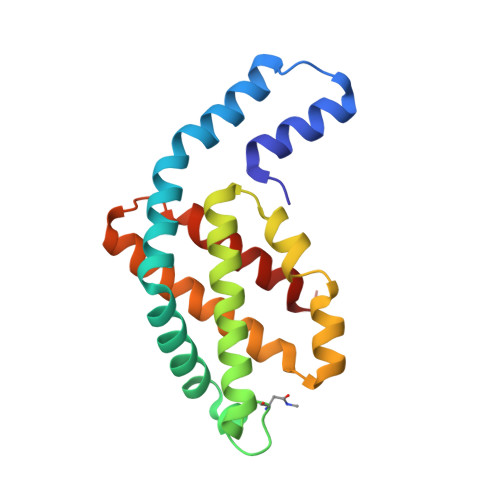
Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome.
Kawakami, K., Hamaguchi, T., Hirose, Y., Kosumi, D., Miyata, M., Kamiya, N., Yonekura, K.(2022) Nat Commun 13: 3389-3389
- PubMed: 35715389
- DOI: https://doi.org/10.1038/s41467-022-30962-9
- Primary Citation of Related Structures:
7VEA, 7VEB - PubMed Abstract:
Cyanobacteria, glaucophytes, and rhodophytes utilize giant, light-harvesting phycobilisomes (PBSs) for capturing solar energy and conveying it to photosynthetic reaction centers. PBSs are compositionally and structurally diverse, and exceedingly complex, all of which pose a challenge for a comprehensive understanding of their function. To date, three detailed architectures of PBSs by cryo-electron microscopy (cryo-EM) have been described: a hemiellipsoidal type, a block-type from rhodophytes, and a cyanobacterial hemidiscoidal-type. Here, we report cryo-EM structures of a pentacylindrical allophycocyanin core and phycocyanin-containing rod of a thermophilic cyanobacterial hemidiscoidal PBS. The structures define the spatial arrangement of protein subunits and chromophores, crucial for deciphering the energy transfer mechanism. They reveal how the pentacylindrical core is formed, identify key interactions between linker proteins and the bilin chromophores, and indicate pathways for unidirectional energy transfer.
Organizational Affiliation:
Biostructural Mechanism Laboratory, RIKEN SPring-8 Center, 1-1-1, Sayo, Hyogo, 679-5148, Japan. kawakami.k@spring8.or.jp.