Structural Basis of the Immunity Mechanisms of Pediocin-like Bacteriocins.
Zhu, L., Zeng, J., Wang, J.(2022) Appl Environ Microbiol 88: e0048122-e0048122
- PubMed: 35703550
- DOI: https://doi.org/10.1128/aem.00481-22
- Primary Citation of Related Structures:
7XNO, 7XTG - PubMed Abstract:
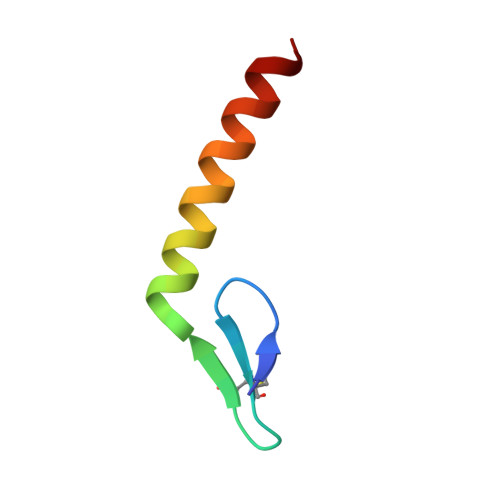
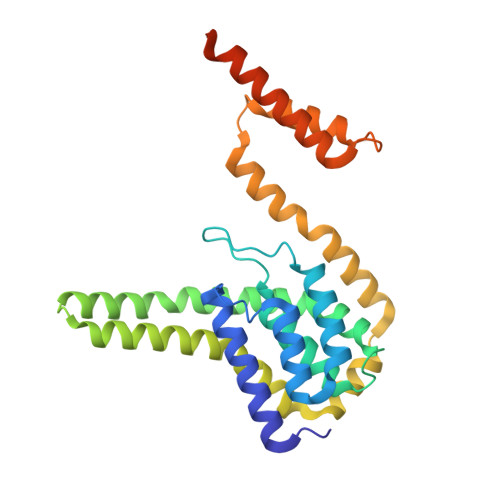
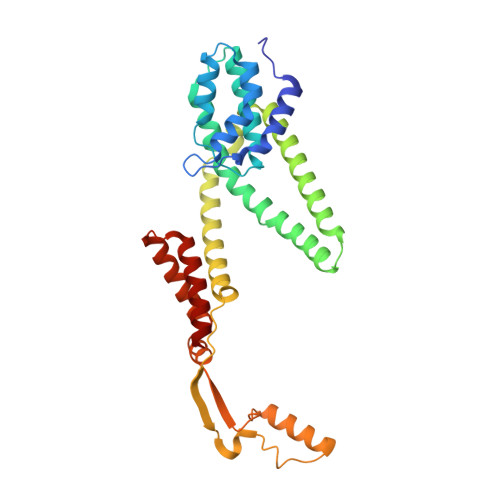
Pediocin-like bacteriocins, also designated class IIa bacteriocins, are ribosomally synthesized antimicrobial peptides targeting species closely related to the producers. They act on the cytoplasmic membrane of Gram-positive cells by dissipating the transmembrane electrical potential through pore formation with the mannose phosphotransferase system (man-PTS) as the target/receptor. Bacteriocin-producing strains also synthesize a cognate immunity protein that protects them against their own bacteriocins. Herein, we report the cryo-electron microscopy structure of the bacteriocin-receptor-immunity ternary complex from Lactobacillus sakei. The complex structure reveals that pediocin-like bacteriocins bind to the same position on the Core domain of man-PTS, while the C-terminal helical tails of bacteriocins delimit the opening range of the Core domain away from the Vmotif domain to facilitate transmembrane pore formation. Upon attack of bacteriocins from the extracellular side, man-PTS exposes its cytosolic side for recognition of the N-terminal four-helix bundle of the immunity protein. The C-terminal loop of the immunity protein then inserts into the pore and blocks leakage induced by bacteriocins. Elucidation of the toxicity and immunity mechanisms of pediocin-like bacteriocins could support the design of novel bacteriocins against antibiotic-resistant pathogenic bacteria. IMPORTANCE Pediocin-like bacteriocins, ribosomally synthesized antimicrobial peptides, are generally co-expressed with cognate immunity proteins to protect the bacteriocin-producing strain from its own bacteriocin. Bacteriocins are considered potential alternatives to conventional antibiotics in the context of the bacterial resistance crisis, but the immunity mechanism is unclear. This study uncovered the mechanisms of action and immunity of class IIa bacteriocins.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, P. R. China.