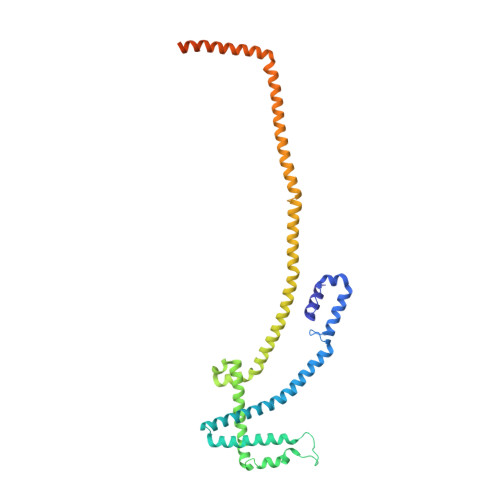
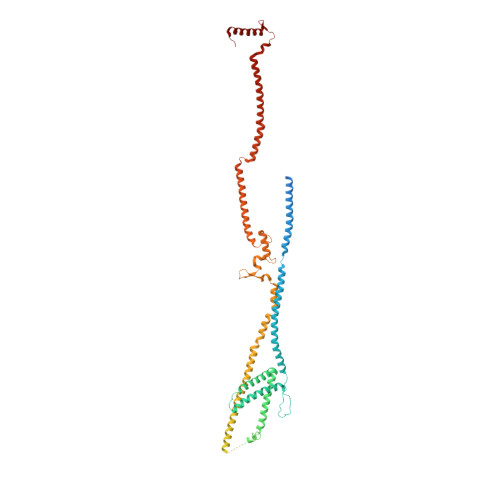
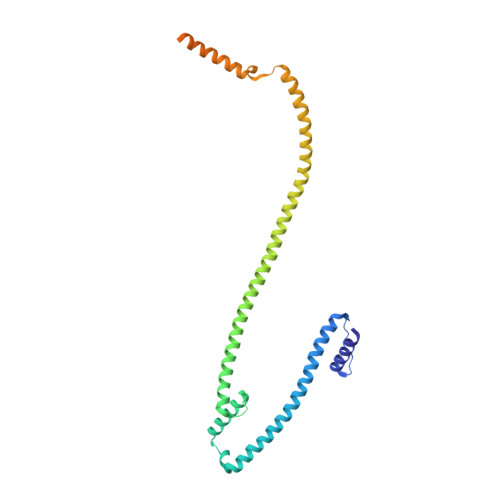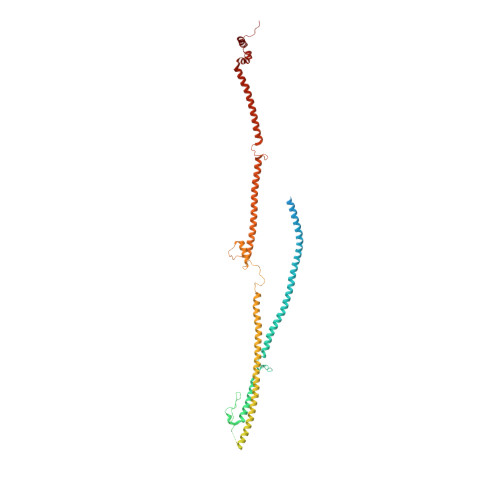The augmin complex architecture reveals structural insights into microtubule branching.
Zupa, E., Wurtz, M., Neuner, A., Hoffmann, T., Rettel, M., Bohler, A., Vermeulen, B.J.A., Eustermann, S., Schiebel, E., Pfeffer, S.(2022) Nat Commun 13: 5635-5635
- PubMed: 36163468
- DOI: https://doi.org/10.1038/s41467-022-33228-6
- Primary Citation of Related Structures:
8AT2, 8AT3, 8AT4 - PubMed Abstract:
In mitosis, the augmin complex binds to spindle microtubules to recruit the γ-tubulin ring complex (γ-TuRC), the principal microtubule nucleator, for the formation of branched microtubules. Our understanding of augmin-mediated microtubule branching is hampered by the lack of structural information on the augmin complex. Here, we elucidate the molecular architecture and conformational plasticity of the augmin complex using an integrative structural biology approach. The elongated structure of the augmin complex is characterised by extensive coiled-coil segments and comprises two structural elements with distinct but complementary functions in γ-TuRC and microtubule binding, linked by a flexible hinge. The augmin complex is recruited to microtubules via a composite microtubule binding site comprising a positively charged unordered extension and two calponin homology domains. Our study provides the structural basis for augmin function in branched microtubule formation, decisively fostering our understanding of spindle formation in mitosis.
Organizational Affiliation:
Zentrum für Molekulare Biologie der Universität Heidelberg, DKFZ-ZMBH Allianz, Im Neuenheimer Feld 282, 69120, Heidelberg, Germany.