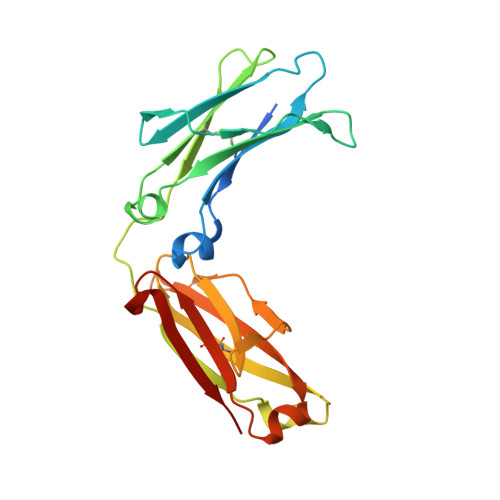
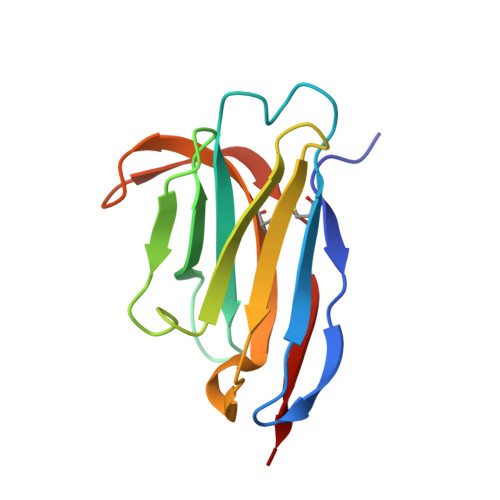
Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Gupta, A., Kao, K.S., Yamin, R., Oren, D.A., Goldgur, Y., Du, J., Lollar, P., Sundberg, E.J., Ravetch, J.V.(2023) Nat Commun 14: 2853-2853
- PubMed: 37202422
- DOI: https://doi.org/10.1038/s41467-023-38453-1
- Primary Citation of Related Structures:
8F8V, 8F8W, 8F8X - PubMed Abstract:
Immunoglobulin G (IgG) antibodies contain a complex N-glycan embedded in the hydrophobic pocket between its heavy chain protomers. This glycan contributes to the structural organization of the Fc domain and determines its specificity for Fcγ receptors, thereby dictating distinct cellular responses. The variable construction of this glycan structure leads to highly-related, but non-equivalent glycoproteins known as glycoforms. We previously reported synthetic nanobodies that distinguish IgG glycoforms. Here, we present the structure of one such nanobody, X0, in complex with the Fc fragment of afucosylated IgG1. Upon binding, the elongated CDR3 loop of X0 undergoes a conformational shift to access the buried N-glycan and acts as a 'glycan sensor', forming hydrogen bonds with the afucosylated IgG N-glycan that would otherwise be sterically hindered by the presence of a core fucose residue. Based on this structure, we designed X0 fusion constructs that disrupt pathogenic afucosylated IgG1-FcγRIIIa interactions and rescue mice in a model of dengue virus infection.
Organizational Affiliation:
Laboratory of Molecular Genetics & Immunology, The Rockefeller University, New York, NY, USA.