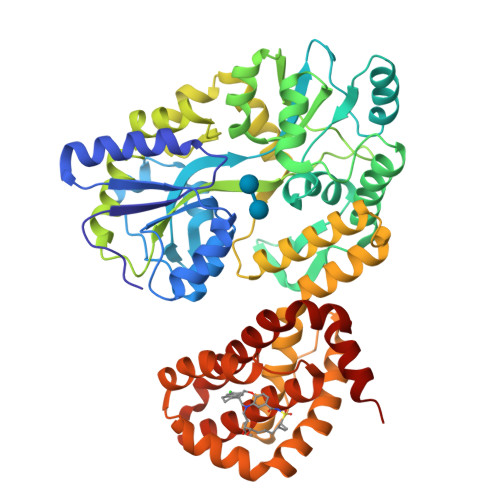
Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
Romanov-Michailidis, F., Hsiao, C.C., Urner, L.M., Jerhaoui, S., Surkyn, M., Miller, B., Vos, A., Dominguez Blanco, M., Bueters, R., Vinken, P., Bekkers, M., Walker, D., Pietrak, B., Eyckmans, W., Dores-Sousa, J.L., Joo Koo, S., Lento, W., Bauser, M., Philippar, U., Rombouts, F.J.R.(2023) J Med Chem 66: 6122-6148
- PubMed: 37114951
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01953
- Primary Citation of Related Structures:
8G3S, 8G3T, 8G3U, 8G3W, 8G3X, 8G3Y - PubMed Abstract:
Avoidance of apoptosis is critical for the development and sustained growth of tumors. The pro-survival protein myeloid cell leukemia 1 (Mcl-1) is an anti-apoptotic member of the Bcl-2 family of proteins which is overexpressed in many cancers. Upregulation of Mcl-1 in human cancers is associated with high tumor grade, poor survival, and resistance to chemotherapy. Therefore, pharmacological inhibition of Mcl-1 is regarded as an attractive approach to treating relapsed or refractory malignancies. Herein, we disclose the design, synthesis, optimization, and early preclinical evaluation of a potent and selective small-molecule inhibitor of Mcl-1. Our exploratory design tactics focused on structural modifications which improve the potency and physicochemical properties of the inhibitor while minimizing the risk of functional cardiotoxicity. Despite being in the "non-Lipinski" beyond-Rule-of-Five property space, the developed compound benefits from exquisite oral bioavailability in vivo and induces potent pharmacodynamic inhibition of Mcl-1 in a mouse xenograft model.
Organizational Affiliation:
Janssen Pharmaceutica NV, Turnhoutseweg 30, B-2340 Beerse, Belgium.