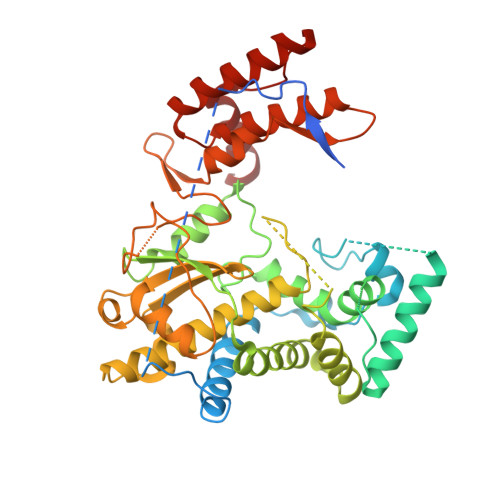
Unusual substructure conformations observed in crystal structures of a dicistrovirus RNA-dependent RNA polymerase suggest contribution of the N-terminal extension in proper folding.
Fang, X., Lu, G., Deng, Y., Yang, S., Hou, C., Gong, P.(2023) Virol Sin 38: 531-540
- PubMed: 37156298
- DOI: https://doi.org/10.1016/j.virs.2023.05.002
- Primary Citation of Related Structures:
8IIB, 8IIC - PubMed Abstract:
The Dicistroviridae is a virus family that includes many insect pathogens. These viruses contain a positive-sense RNA genome that is replicated by the virally encoded RNA-dependent RNA polymerase (RdRP) also named 3D pol . Compared with the Picornaviridae RdRPs such as poliovirus (PV) 3D pol , the Dicistroviridae representative Israeli acute paralysis virus (IAPV) 3D pol has an additional N-terminal extension (NE) region that is about 40-residue in length. To date, both the structure and catalytic mechanism of the Dicistroviridae RdRP have remain elusive. Here we reported crystal structures of two truncated forms of IAPV 3D pol , namely Δ85 and Δ40, both missing the NE region, and the 3D pol protein in these structures exhibited three conformational states. The palm and thumb domains of these IAPV 3D pol structures are largely consistent with those of the PV 3D pol structures. However, in all structures, the RdRP fingers domain is partially disordered, while different conformations of RdRP substructures and interactions between them are also present. In particular, a large-scale conformational change occurred in the motif B-middle finger region in one protein chain of the Δ40 structure, while a previously documented alternative conformation of motif A was observed in all IAPV structures. These experimental data on one hand show intrinsic conformational variances of RdRP substructures, and on the other hand suggest possible contribution of the NE region in proper RdRP folding in IAPV.
Organizational Affiliation:
Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, 430207, China; University of Chinese Academy of Sciences, Beijing, 100049, China.