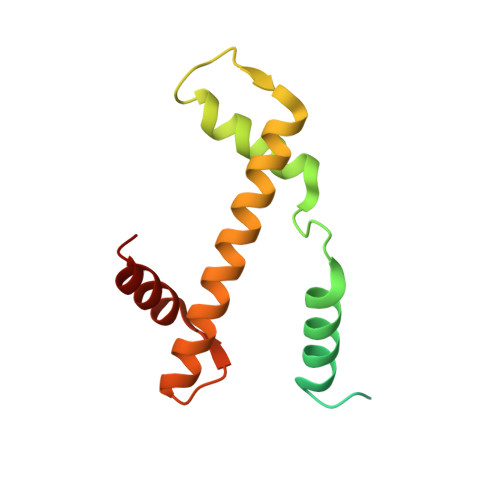
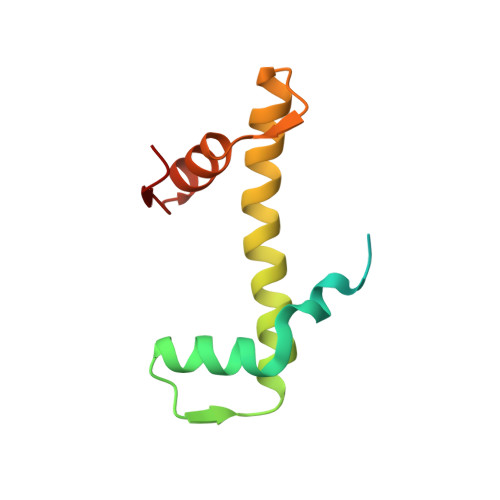
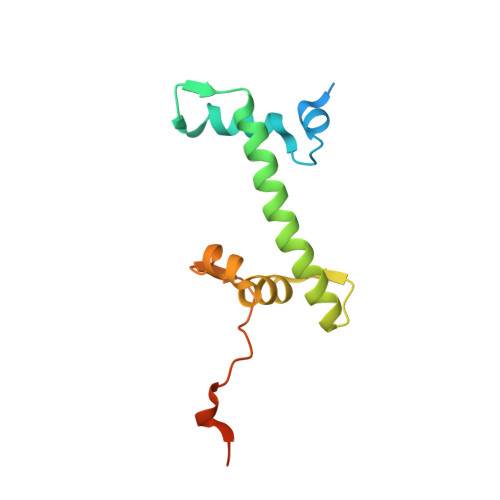
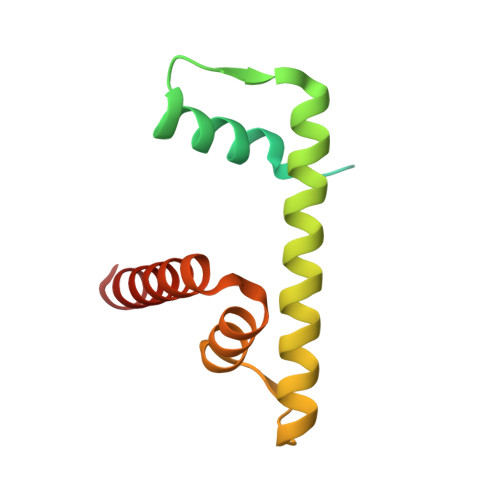
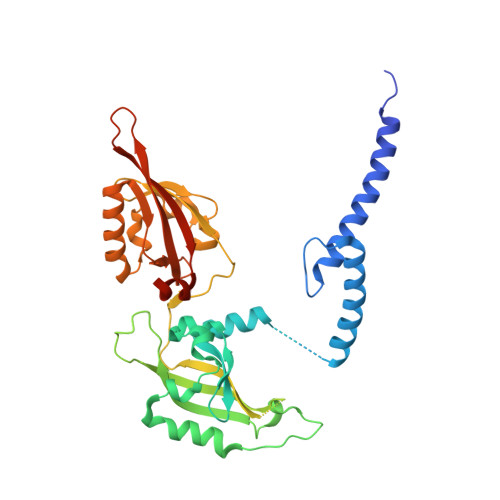
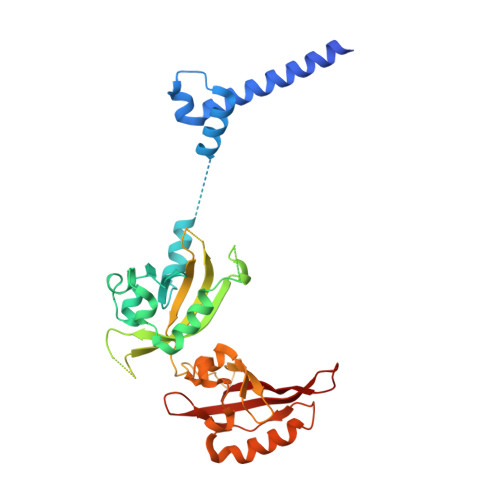
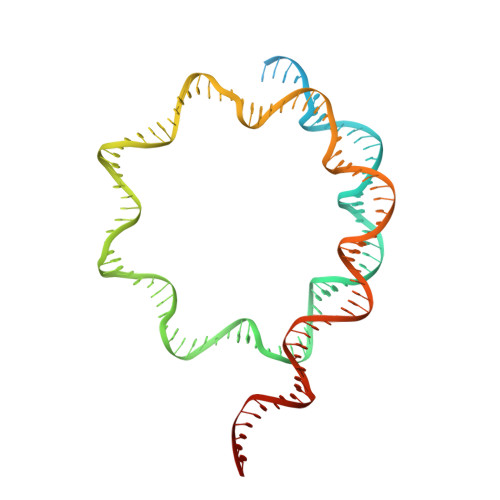
Cooperation between bHLH transcription factors and histones for DNA access.
Michael, A.K., Stoos, L., Crosby, P., Eggers, N., Nie, X.Y., Makasheva, K., Minnich, M., Healy, K.L., Weiss, J., Kempf, G., Cavadini, S., Kater, L., Seebacher, J., Vecchia, L., Chakraborty, D., Isbel, L., Grand, R.S., Andersch, F., Fribourgh, J.L., Schubeler, D., Zuber, J., Liu, A.C., Becker, P.B., Fierz, B., Partch, C.L., Menet, J.S., Thoma, N.H.(2023) Nature 619: 385-393
- PubMed: 37407816
- DOI: https://doi.org/10.1038/s41586-023-06282-3
- Primary Citation of Related Structures:
8OSJ, 8OSK, 8OSL, 8OTS, 8OTT, 9A3O, 9A3P - PubMed Abstract:
The basic helix-loop-helix (bHLH) family of transcription factors recognizes DNA motifs known as E-boxes (CANNTG) and includes 108 members 1 . Here we investigate how chromatinized E-boxes are engaged by two structurally diverse bHLH proteins: the proto-oncogene MYC-MAX and the circadian transcription factor CLOCK-BMAL1 (refs. 2,3 ). Both transcription factors bind to E-boxes preferentially near the nucleosomal entry-exit sites. Structural studies with engineered or native nucleosome sequences show that MYC-MAX or CLOCK-BMAL1 triggers the release of DNA from histones to gain access. Atop the H2A-H2B acidic patch 4 , the CLOCK-BMAL1 Per-Arnt-Sim (PAS) dimerization domains engage the histone octamer disc. Binding of tandem E-boxes 5-7 at endogenous DNA sequences occurs through direct interactions between two CLOCK-BMAL1 protomers and histones and is important for circadian cycling. At internal E-boxes, the MYC-MAX leucine zipper can also interact with histones H2B and H3, and its binding is indirectly enhanced by OCT4 elsewhere on the nucleosome. The nucleosomal E-box position and the type of bHLH dimerization domain jointly determine the histone contact, the affinity and the degree of competition and cooperativity with other nucleosome-bound factors.
- Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
Organizational Affiliation: